This page was generated from
docs/Examples/Feldspar_Thermobarometry/Fspar_Liq_Matching.ipynb.
Interactive online version:
.
Plag-Liq and Kspar-Liq Matching
This notebook shows how to assess all possible matches for Plag-Liq and Kspar-Liq pairs where there aren’t obvious pairwise analyses
You can download the excel spreadsheet here: https://github.com/PennyWieser/Thermobar/blob/main/docs/Examples/Feldspar_Thermobarometry/Feldspar_Liquid.xlsx
[1]:
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import Thermobar as pt
pd.options.display.max_columns = None
Example 1 - Plag and Liquid.
[2]:
## Load in the liquids
out_Liq=pt.import_excel('Feldspar_Liquid.xlsx', sheet_name="Liq_only")
Liqs=out_Liq['Liqs']
# Load in the Plags
out_Plag=pt.import_excel('Feldspar_Liquid.xlsx', sheet_name="Plag_only")
Plags=out_Plag['Plags']
[3]:
## Inspect them to check they read in right
display(Plags.head())
display(Liqs.head())
| SiO2_Plag | TiO2_Plag | Al2O3_Plag | FeOt_Plag | MnO_Plag | MgO_Plag | CaO_Plag | Na2O_Plag | K2O_Plag | Cr2O3_Plag | Sample_ID_Plag | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 57.3 | 0.09 | 26.6 | 0.43 | 0.0 | 0.03 | 8.33 | 6.11 | 0.49 | 0.0 | Plag1 |
| 1 | 56.5 | 0.12 | 26.9 | 0.47 | 0.0 | 0.05 | 8.95 | 5.66 | 0.47 | 0.0 | Plag2 |
| 2 | 57.6 | 0.11 | 26.3 | 0.50 | 0.0 | 0.07 | 8.50 | 6.27 | 0.40 | 0.0 | Plag3_core |
| 3 | 57.2 | 0.16 | 27.0 | 0.62 | 0.0 | 0.06 | 9.03 | 5.58 | 0.84 | 0.0 | Plag4 |
| 4 | 56.7 | 0.14 | 27.6 | 0.69 | 0.0 | 0.11 | 9.46 | 5.58 | 0.48 | 0.0 | Plag5 |
| SiO2_Liq | TiO2_Liq | Al2O3_Liq | FeOt_Liq | MnO_Liq | MgO_Liq | CaO_Liq | Na2O_Liq | K2O_Liq | Cr2O3_Liq | P2O5_Liq | H2O_Liq | Fe3Fet_Liq | NiO_Liq | CoO_Liq | CO2_Liq | Sample_ID_Liq | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 49.1 | 3.22 | 14.4 | 14.8 | 0.14 | 3.20 | 6.72 | 3.34 | 1.70 | 0.0 | 1.13 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | Glass1 |
| 1 | 49.2 | 3.89 | 15.3 | 13.7 | 0.12 | 3.88 | 6.76 | 3.44 | 1.22 | 0.0 | 0.83 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | Glass2 |
| 2 | 49.6 | 3.79 | 15.8 | 13.0 | 0.14 | 4.26 | 6.59 | 3.65 | 1.04 | 0.0 | 0.63 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | Glass3 |
| 3 | 47.1 | 4.21 | 12.0 | 17.8 | 0.18 | 3.40 | 7.28 | 2.93 | 2.02 | 0.0 | 2.32 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | Glass4 |
| 4 | 48.1 | 3.88 | 13.2 | 16.4 | 0.16 | 4.02 | 6.51 | 3.36 | 1.36 | 0.0 | 1.59 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | Glass5 |
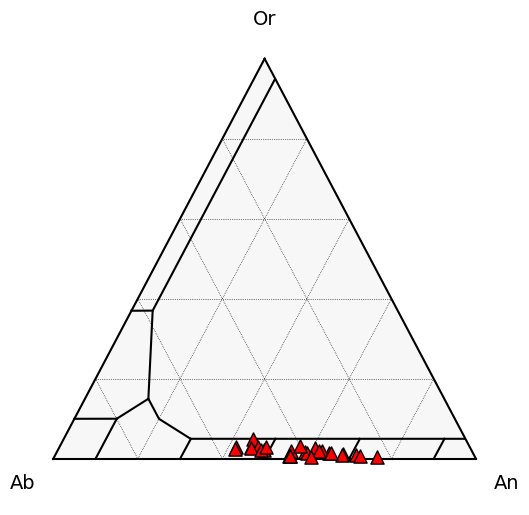
Please check your feldspars are actually plagioclases! If you apply plag-liq thermometers to kspars, you get stupid results, that mess up the iterative calculations
Lets plot to inspect and check they are actually plag, else you will have convergence issues
-This function relies heavily on the ternary plot package from Marc Harper et al. 2015 - https://github.com/marcharper/python-ternary, if you use these figures, you must cite that (Marc Harper et al. (2015). python-ternary: Ternary Plots in Python. Zenodo. 10.5281/zenodo.594435) as well as Thermobar.
You may have problems with this package if you have the separate “ternary” package installed (yes, there are python packages called ternary and python-ternary- Yay!). I (penny) got the error “module ternary has no attribute figure, so had to uninstall the ternary I had through pip (pip uninstall ternary), and re-install python-ternary through conda in the command line “conda install python-ternary”. If you have everything in pip, or conda, keep in 1 environment, don’t follow my bad example here!
[4]:
Plag_tern_points = pt.tern_points_fspar(fspar_comps=Plags)
fig, tax = pt.plot_fspar_classification(major_grid=True, ticks=False)
# Here we plot the data ontop, with plag as red triangles
tax.scatter(
Plag_tern_points,
edgecolor="k",
marker="^",
facecolor="red",
label='Plag',
s=90
)
[4]:
<AxesSubplot:>
[5]:
Plag_components=pt.calculate_cat_fractions_plagioclase(plag_comps=Plags)
RealPlags=Plags.loc[Plag_components['An_Plag']>0.05].reset_index(drop=True)
Example 1a - All possible Liq-Plag matches, An-Ab equilibrium
You can specify to use the An-Ab equilibrium test of Putirka (2008) as in this example
[6]:
MM_dict=pt.calculate_fspar_liq_temp_matching(liq_comps=Liqs, plag_comps=RealPlags,
equationT="T_Put2008_eq24a", P=5,
Ab_An_P2008=True, H2O_Liq=2)
Av_Matches=MM_dict['Av_PTs']
All_Matches=MM_dict['All_PTs']
Considering N=28 Fspar & N=24 Liqs, which is a total of N=672 Liq-Fspar pairs, be patient if this is >>1 million!
Applying filter to only average those that pass the An-Ab eq test of Putirka, 2008
Done!!! I found a total of N=327 Fspar-Liq matches using the specified filter. N=28 Fspar out of the N=28 Fspar that you input matched to 1 or more liquids
Example 1b - All possible Liq-Plag matches, no filter
This could be used if you want to develop your own equilibrium filters
[7]:
MM_dict=pt.calculate_fspar_liq_temp_matching(liq_comps=Liqs, plag_comps=RealPlags,
equationT="T_Put2008_eq24a", P=5,
Ab_An_P2008=False, H2O_Liq=2)
Av_Matches=MM_dict['Av_PTs']
All_Matches=MM_dict['All_PTs']
Considering N=28 Fspar & N=24 Liqs, which is a total of N=672 Liq-Fspar pairs, be patient if this is >>1 million!
We are returning all pairs, if you want to use the Ab-An equilibrium test of Putirka (2008), enter Ab_An_P2008=True
Done!!! I found a total of N=672 Fspar-Liq matches using the specified filter. N=28 Fspar out of the N=28 Fspar that you input matched to 1 or more liquids
Example 1c - Iterating T and H2O for all possible Plag-Liq matches
[8]:
T_H_Iter_dict=pt.calculate_fspar_liq_temp_hygr_matching(liq_comps=Liqs, plag_comps=RealPlags,
equationT="T_Put2008_eq24a", equationH="H_Waters2015", P=5)
T_H_Calc_Av=T_H_Iter_dict.get('Av_HTs')
T_H_Calc_All=T_H_Iter_dict.get('All_HTs')
T_H_Evol=T_H_Iter_dict.get('T_H_Evolution')
Considering N=28 Fspar & N=24 Liqs, which is a total of N=672 Liq- Fspar pairs, be patient if this is >>1 million!
100%|██████████████████████████████████████████████████████████████████████████████████| 20/20 [00:02<00:00, 9.58it/s]
Applying filter to only average those that pass the An-Ab eq test of Putirka, 2008
Done!!! I found a total of N=327 Fspar-Liq matches using the specified filter. N=28 Fspar out of the N=28 Fspar that you input matched to 1 or more liquids
[9]:
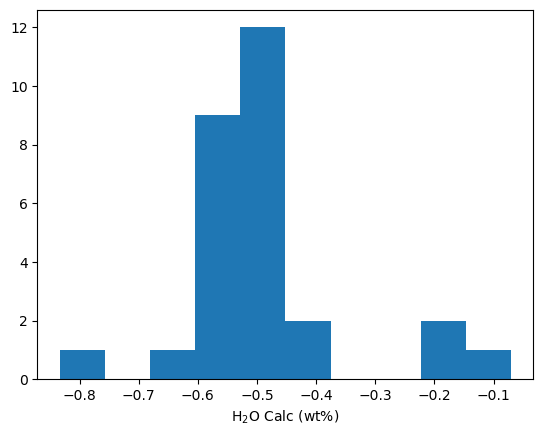
# Maybe you want to plot a histogram of H2O contents
plt.hist(T_H_Calc_Av['Mean_H2O_calc'])
plt.xlabel('H$_2$O Calc (wt%)')
[9]:
Text(0.5, 0, 'H$_2$O Calc (wt%)')
Example 2 - Kspar-Liquid calculations
[10]:
# Load in the Kspars
out_Kspar=pt.import_excel('Feldspar_Liquid.xlsx', sheet_name="Kspar_only")
Kspars=out_Kspar['Kspars']
# Load in the liquids you want to match with
out_Liq2=pt.import_excel('Feldspar_Liquid.xlsx', sheet_name="Evolved_Liq_only")
Liqs=out_Liq2['Liqs']
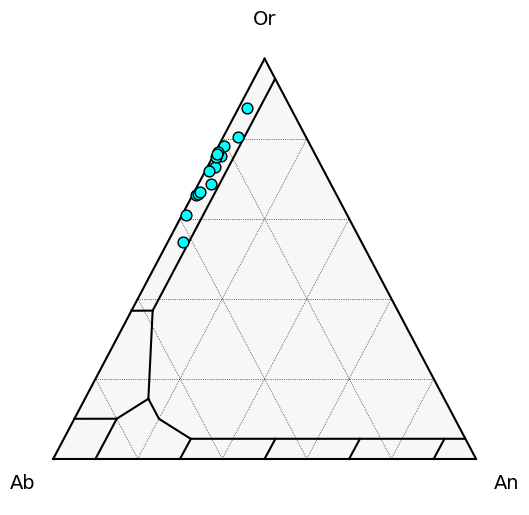
Check actually kspar
[11]:
Kspar_tern_points = pt.tern_points_fspar(fspar_comps=Kspars)
fig, tax = pt.plot_fspar_classification(major_grid=True, ticks=False)
tax.scatter(
Kspar_tern_points,
edgecolor="k",
marker="o",
facecolor="cyan",
s=60, label='Kspar'
)
[11]:
<AxesSubplot:>
[12]:
Kspar_components=pt.calculate_cat_fractions_kspar(kspar_comps=Kspars)
RealKspars=Kspars.loc[Kspar_components['An_Kspar']<0.05].reset_index(drop=True)
2a - Lets calculate temperature for all liq-kspar matches
Currently no equilibrium tests exist for Kspar-Liquid that we are aware off. If you know of ones I can implement, please reach out!
[15]:
melt_match_2=pt.calculate_fspar_liq_temp_matching(liq_comps=Liqs, kspar_comps=RealKspars,
equationT="T_Put2008_eq24b", P=5,
Ab_An_P2008=True, H2O_Liq=2)
melt_match_2['Av_PTs'].head()
Considering N=15 Fspar & N=15 Liqs, which is a total of N=225 Liq-Fspar pairs, be patient if this is >>1 million!
Sorry, no equilibrium tests implemented for Kspar-Liquid. Weve returned all possible pairs, you will have to filter them yourselves
Done!!! I found a total of N=225 Fspar-Liq matches using the specified filter. N=15 Fspar out of the N=15 Fspar that you input matched to 1 or more liquids
[15]:
| ID_Fspar | Mean_Sample_ID_Kspar | Mean_T_K_calc | Mean_SiO2_Liq | Mean_TiO2_Liq | Mean_Al2O3_Liq | Mean_FeOt_Liq | Mean_MnO_Liq | Mean_MgO_Liq | Mean_CaO_Liq | Mean_Na2O_Liq | Mean_K2O_Liq | Mean_Cr2O3_Liq | Mean_P2O5_Liq | Mean_H2O_Liq | Mean_Fe3Fet_Liq | Mean_NiO_Liq | Mean_CoO_Liq | Mean_CO2_Liq | Mean_Sample_ID_Liq | Mean_SiO2_Liq_mol_frac | Mean_MgO_Liq_mol_frac | Mean_MnO_Liq_mol_frac | Mean_FeOt_Liq_mol_frac | Mean_CaO_Liq_mol_frac | Mean_Al2O3_Liq_mol_frac | Mean_Na2O_Liq_mol_frac | Mean_K2O_Liq_mol_frac | Mean_TiO2_Liq_mol_frac | Mean_P2O5_Liq_mol_frac | Mean_Cr2O3_Liq_mol_frac | Mean_Si_Liq_cat_frac | Mean_Mg_Liq_cat_frac | Mean_Mn_Liq_cat_frac | Mean_Fet_Liq_cat_frac | Mean_Ca_Liq_cat_frac | Mean_Al_Liq_cat_frac | Mean_Na_Liq_cat_frac | Mean_K_Liq_cat_frac | Mean_Ti_Liq_cat_frac | Mean_P_Liq_cat_frac | Mean_Cr_Liq_cat_frac | Mean_Mg_Number_Liq_NoFe3 | Mean_Mg_Number_Liq_Fe3 | Mean_ID_liq | Mean_Sample_ID_liq | # of Liqs Averaged | Std_Sample_ID_Kspar | Std_T_K_calc | Std_SiO2_Liq | Std_TiO2_Liq | Std_Al2O3_Liq | Std_FeOt_Liq | Std_MnO_Liq | Std_MgO_Liq | Std_CaO_Liq | Std_Na2O_Liq | Std_K2O_Liq | Std_Cr2O3_Liq | Std_P2O5_Liq | Std_H2O_Liq | Std_Fe3Fet_Liq | Std_NiO_Liq | Std_CoO_Liq | Std_CO2_Liq | Std_Sample_ID_Liq | Std_SiO2_Liq_mol_frac | Std_MgO_Liq_mol_frac | Std_MnO_Liq_mol_frac | Std_FeOt_Liq_mol_frac | Std_CaO_Liq_mol_frac | Std_Al2O3_Liq_mol_frac | Std_Na2O_Liq_mol_frac | Std_K2O_Liq_mol_frac | Std_TiO2_Liq_mol_frac | Std_P2O5_Liq_mol_frac | Std_Cr2O3_Liq_mol_frac | Std_Si_Liq_cat_frac | Std_Mg_Liq_cat_frac | Std_Mn_Liq_cat_frac | Std_Fet_Liq_cat_frac | Std_Ca_Liq_cat_frac | Std_Al_Liq_cat_frac | Std_Na_Liq_cat_frac | Std_K_Liq_cat_frac | Std_Ti_Liq_cat_frac | Std_P_Liq_cat_frac | Std_Cr_Liq_cat_frac | Std_Mg_Number_Liq_NoFe3 | Std_Mg_Number_Liq_Fe3 | Std_ID_liq | Std_Sample_ID_liq | Sample_ID_Kspar | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | Kspar1 | 1138.165453 | 62.976667 | 0.45 | 18.56 | 3.17 | 0.27 | 0.23 | 1.64 | 5.71 | 6.223333 | 0.0 | 0.02 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 7.0 | 0.709623 | 0.003864 | 0.002577 | 0.029872 | 0.0198 | 0.123242 | 0.062378 | 0.044734 | 0.003814 | 0.000095 | 0.0 | 0.576767 | 0.00314 | 0.002094 | 0.024278 | 0.016092 | 0.200325 | 0.101348 | 0.072699 | 0.0031 | 0.000155 | 0.0 | 0.114521 | 0.114521 | 7.0 | 7.0 | 15 | Kspar1 | 27.575211 | 0.703732 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.507093 | 0.351866 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.472136 | 0.007112 | 0.000006 | 0.000004 | 0.000045 | 0.00003 | 0.000186 | 0.005578 | 0.002604 | 0.000006 | 1.436252e-07 | 0.0 | 0.009076 | 0.000015 | 0.00001 | 0.000115 | 0.000076 | 0.000947 | 0.008549 | 0.003937 | 0.000015 | 7.329643e-07 | 0.0 | 0.0 | 0.0 | 4.472136 | 4.472136 | Kspar1 |
| 1 | 1 | Kspar2 | 1087.464362 | 62.976667 | 0.45 | 18.56 | 3.17 | 0.27 | 0.23 | 1.64 | 5.71 | 6.223333 | 0.0 | 0.02 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 7.0 | 0.709623 | 0.003864 | 0.002577 | 0.029872 | 0.0198 | 0.123242 | 0.062378 | 0.044734 | 0.003814 | 0.000095 | 0.0 | 0.576767 | 0.00314 | 0.002094 | 0.024278 | 0.016092 | 0.200325 | 0.101348 | 0.072699 | 0.0031 | 0.000155 | 0.0 | 0.114521 | 0.114521 | 7.0 | 7.0 | 15 | Kspar2 | 25.122349 | 0.703732 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.507093 | 0.351866 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.472136 | 0.007112 | 0.000006 | 0.000004 | 0.000045 | 0.00003 | 0.000186 | 0.005578 | 0.002604 | 0.000006 | 1.436252e-07 | 0.0 | 0.009076 | 0.000015 | 0.00001 | 0.000115 | 0.000076 | 0.000947 | 0.008549 | 0.003937 | 0.000015 | 7.329643e-07 | 0.0 | 0.0 | 0.0 | 4.472136 | 4.472136 | Kspar2 |
| 2 | 2 | Kspar3 | 977.039695 | 62.976667 | 0.45 | 18.56 | 3.17 | 0.27 | 0.23 | 1.64 | 5.71 | 6.223333 | 0.0 | 0.02 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 7.0 | 0.709623 | 0.003864 | 0.002577 | 0.029872 | 0.0198 | 0.123242 | 0.062378 | 0.044734 | 0.003814 | 0.000095 | 0.0 | 0.576767 | 0.00314 | 0.002094 | 0.024278 | 0.016092 | 0.200325 | 0.101348 | 0.072699 | 0.0031 | 0.000155 | 0.0 | 0.114521 | 0.114521 | 7.0 | 7.0 | 15 | Kspar3 | 20.190486 | 0.703732 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.507093 | 0.351866 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.472136 | 0.007112 | 0.000006 | 0.000004 | 0.000045 | 0.00003 | 0.000186 | 0.005578 | 0.002604 | 0.000006 | 1.436252e-07 | 0.0 | 0.009076 | 0.000015 | 0.00001 | 0.000115 | 0.000076 | 0.000947 | 0.008549 | 0.003937 | 0.000015 | 7.329643e-07 | 0.0 | 0.0 | 0.0 | 4.472136 | 4.472136 | Kspar3 |
| 3 | 3 | Kspar4 | 1023.046234 | 62.976667 | 0.45 | 18.56 | 3.17 | 0.27 | 0.23 | 1.64 | 5.71 | 6.223333 | 0.0 | 0.02 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 7.0 | 0.709623 | 0.003864 | 0.002577 | 0.029872 | 0.0198 | 0.123242 | 0.062378 | 0.044734 | 0.003814 | 0.000095 | 0.0 | 0.576767 | 0.00314 | 0.002094 | 0.024278 | 0.016092 | 0.200325 | 0.101348 | 0.072699 | 0.0031 | 0.000155 | 0.0 | 0.114521 | 0.114521 | 7.0 | 7.0 | 15 | Kspar4 | 22.177217 | 0.703732 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.507093 | 0.351866 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.472136 | 0.007112 | 0.000006 | 0.000004 | 0.000045 | 0.00003 | 0.000186 | 0.005578 | 0.002604 | 0.000006 | 1.436252e-07 | 0.0 | 0.009076 | 0.000015 | 0.00001 | 0.000115 | 0.000076 | 0.000947 | 0.008549 | 0.003937 | 0.000015 | 7.329643e-07 | 0.0 | 0.0 | 0.0 | 4.472136 | 4.472136 | Kspar4 |
| 4 | 4 | Kspar5 | 1073.413522 | 62.976667 | 0.45 | 18.56 | 3.17 | 0.27 | 0.23 | 1.64 | 5.71 | 6.223333 | 0.0 | 0.02 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 7.0 | 0.709623 | 0.003864 | 0.002577 | 0.029872 | 0.0198 | 0.123242 | 0.062378 | 0.044734 | 0.003814 | 0.000095 | 0.0 | 0.576767 | 0.00314 | 0.002094 | 0.024278 | 0.016092 | 0.200325 | 0.101348 | 0.072699 | 0.0031 | 0.000155 | 0.0 | 0.114521 | 0.114521 | 7.0 | 7.0 | 15 | Kspar5 | 24.463649 | 0.703732 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.507093 | 0.351866 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.472136 | 0.007112 | 0.000006 | 0.000004 | 0.000045 | 0.00003 | 0.000186 | 0.005578 | 0.002604 | 0.000006 | 1.436252e-07 | 0.0 | 0.009076 | 0.000015 | 0.00001 | 0.000115 | 0.000076 | 0.000947 | 0.008549 | 0.003937 | 0.000015 | 7.329643e-07 | 0.0 | 0.0 | 0.0 | 4.472136 | 4.472136 | Kspar5 |
[ ]: