This page was generated from
docs/Examples/Liquid_Ol_Liq_Themometry/Olivine_MatrixGlass_Mg_Fe_Eq_SingleSamples.ipynb.
Interactive online version:
.
1 Sample - Fe-Mg equilibrium between olivines and matrix glasses
This notebook shows how to assess Fe-Mg equilibrium between olivines and matrix glases
In this example, we have just 1 sample in our spreadsheet, so we dont have to segment by sample name
You can download the spreadsheet here: https://github.com/PennyWieser/Thermobar/blob/main/docs/Examples/Liquid_Ol_Liq_Themometry/2018_Olivines_Glasses.xlsx
Install Thermobar if you haven’t already (remove the # and press run!)
[1]:
#!pip install Thermobar
[2]:
# Loading various python things
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import Thermobar as pt
Loading a dataset
This dataset is from Wieser et al. (2021) - https://doi.org/10.1029/2020GC009364
It shows olivines and glasses from Kilauea from a single sample LL4
Load in the matrix glasses
[3]:
# We dont have to add _Liq as its all liquids,
# just specify the _Liq in the import
MG_input=pt.import_excel('2018_Olivines_Glasses.xlsx',
sheet_name='LL4_MGs', suffix="_Liq")
MG_all=MG_input['my_input'] ## All columns
MG_Liqs=MG_input['Liqs'] ## Just Liquid columns
MG_Liqs.head()
[3]:
| SiO2_Liq | TiO2_Liq | Al2O3_Liq | FeOt_Liq | MnO_Liq | MgO_Liq | CaO_Liq | Na2O_Liq | K2O_Liq | Cr2O3_Liq | P2O5_Liq | H2O_Liq | Fe3Fet_Liq | NiO_Liq | CoO_Liq | CO2_Liq | Sample_ID_Liq | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 50.6243 | 2.8773 | 13.0360 | 11.8288 | 0.1830 | 5.8471 | 10.2471 | 2.5120 | 0.5234 | 0.0 | 0.2689 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0 |
| 1 | 50.0975 | 2.8840 | 12.8885 | 11.2897 | 0.1769 | 5.8521 | 9.9658 | 2.4681 | 0.5305 | 0.0 | 0.2652 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1 |
| 2 | 50.1346 | 2.8741 | 12.8953 | 11.8696 | 0.1814 | 5.8682 | 10.2012 | 2.4618 | 0.5778 | 0.0 | 0.2875 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2 |
| 3 | 50.5233 | 2.8765 | 12.9818 | 11.3536 | 0.1454 | 5.8673 | 10.1806 | 2.6206 | 0.5206 | 0.0 | 0.2759 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3 |
| 4 | 51.5840 | 2.9055 | 13.3290 | 11.5014 | 0.2286 | 5.8450 | 10.0408 | 2.1722 | 0.5565 | 0.0 | 0.2693 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4 |
Load in the olivines
[4]:
Ols_in=pt.import_excel('2018_Olivines_Glasses.xlsx',
sheet_name='LL4_Ols')
Ols_Ol=Ols_in['Ols']
Ols_input=Ols_in['my_input']
Workflow 1: Assess if the olivines are in equilibrium with the co-erupted matrix glass
1. Calculate Mg# for liquids
[5]:
Liq_Mgno_calc=pt.calculate_liq_mgno(liq_comps=MG_Liqs, Fe3Fet_Liq=0.15)
Liq_Mgno_calc.head()
# Calculate mean value to plot
Liq_Mgno_calc_mean=np.mean(Liq_Mgno_calc)
2. Calculate Olivine Fo contents
[6]:
Ol_Fo_Calc=pt.calculate_ol_fo(ol_comps=Ols_Ol)
3. Calculate fields you want to plot on a Rhodes diagram
[7]:
Rhodes=pt.calculate_ol_rhodes_diagram_lines(Min_Mgno=0.5, Max_Mgno=0.7)
Rhodes.head()
[7]:
| Mg#_Liq | Eq_Ol_Fo_Roeder (Kd=0.3) | Eq_Ol_Fo_Roeder (Kd=0.27) | Eq_Ol_Fo_Roeder (Kd=0.33) | Eq_Ol_Fo_Matzen (Kd=0.34) | Eq_Ol_Fo_Matzen (Kd=0.328) | Eq_Ol_Fo_Matzen (Kd=0.352) | |
|---|---|---|---|---|---|---|---|
| 0 | 0.500000 | 0.769231 | 0.787402 | 0.751880 | 0.746269 | 0.753012 | 0.739645 |
| 1 | 0.502020 | 0.770662 | 0.788751 | 0.753384 | 0.747796 | 0.754512 | 0.741198 |
| 2 | 0.504040 | 0.772087 | 0.790095 | 0.754883 | 0.749317 | 0.756006 | 0.742745 |
| 3 | 0.506061 | 0.773506 | 0.791432 | 0.756375 | 0.750832 | 0.757493 | 0.744286 |
| 4 | 0.508081 | 0.774919 | 0.792763 | 0.757861 | 0.752341 | 0.758975 | 0.745822 |
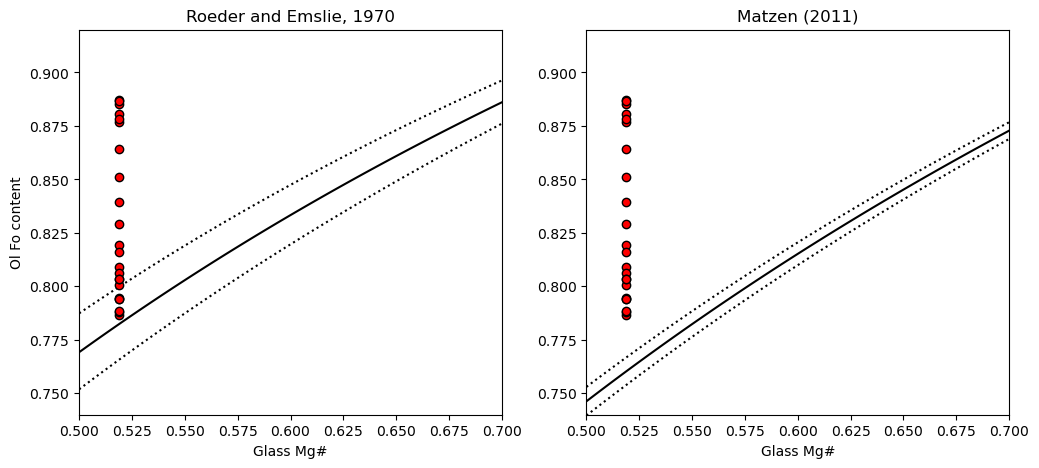
4. Plot the Kd model you want, along with the olivine and glass Mg
[9]:
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(12,5))
# Plotting for Roeder and Emslie
ax1.set_title('Roeder and Emslie, 1970')
# Plotting equilibrium lines
ax1.plot(Rhodes['Mg#_Liq'], Rhodes['Eq_Ol_Fo_Roeder (Kd=0.27)'], ':k')
ax1.plot(Rhodes['Mg#_Liq'], Rhodes['Eq_Ol_Fo_Roeder (Kd=0.33)'], ':k')
ax1.plot(Rhodes['Mg#_Liq'], Rhodes['Eq_Ol_Fo_Roeder (Kd=0.3)'], '-k')
# Plotting data
ax1.plot(Ol_Fo_Calc*0+Liq_Mgno_calc_mean, Ol_Fo_Calc, 'ok', mfc='red')
ax2.set_title('Matzen (2011)')
# Plotting equilibrium lines
ax2.plot(Rhodes['Mg#_Liq'], Rhodes['Eq_Ol_Fo_Matzen (Kd=0.328)'], ':k')
ax2.plot(Rhodes['Mg#_Liq'], Rhodes['Eq_Ol_Fo_Matzen (Kd=0.352)'], ':k')
ax2.plot(Rhodes['Mg#_Liq'], Rhodes['Eq_Ol_Fo_Matzen (Kd=0.34)'], '-k')
# Plotting data
ax2.plot(Ol_Fo_Calc*0+Liq_Mgno_calc_mean, Ol_Fo_Calc, 'ok', mfc='red')
ax1.set_ylabel('Ol Fo content')
ax1.set_xlabel('Glass Mg#')
ax2.set_xlabel('Glass Mg#')
# You may have to adjust axes limits
ax1.set_ylim([0.74, 0.92])
ax2.set_ylim([0.74, 0.92])
ax1.set_xlim([0.5, 0.7])
ax2.set_xlim([0.5, 0.7])
[9]:
(0.5, 0.7)
[ ]:
[ ]:
[ ]: