This page was generated from
docs/Examples/Cpx_Cpx_Liq_Thermobarometry/Cpx_Liquid_melt_matching/Cpx_MeltMatch1_Gleeson2020.ipynb.
Interactive online version:
.
Cpx-Liquid Melt Matching (Simple Intro)
This notebook recreates the method of Gleeson et al. (2020) - JPET - https://doi.org/10.1093/petrology/egaa094
for a more advanced example, see Cpx_MeltMatch2_ScruggsPutirka
You can download the excel file with the data from https://github.com/PennyWieser/Thermobar/blob/main/docs/Examples/Cpx_Cpx_Liq_Thermobarometry/Cpx_Liquid_melt_matching/Gleeson2020JPET_Input_Pyroxene_Melts.xlsx
You need to install Thermobar once on your machine, if you haven’t done this yet, uncomment the line below (remove the #)
[1]:
#!pip install Thermobar
This imports all the python things you need
[1]:
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
!pip install Thermobar
import Thermobar as pt
Requirement already satisfied: Thermobar in g:\my drive\postdoc\pymme\mybarometers\thermobar_outer\src (1.0.10)
Requirement already satisfied: pandas in c:\users\penny\anaconda3\lib\site-packages (from Thermobar) (1.4.4)
Requirement already satisfied: numpy in c:\users\penny\anaconda3\lib\site-packages (from Thermobar) (1.24.2)
Requirement already satisfied: python-ternary in c:\users\penny\anaconda3\lib\site-packages (from Thermobar) (1.0.8)
Requirement already satisfied: matplotlib in c:\users\penny\anaconda3\lib\site-packages (from Thermobar) (3.5.2)
Requirement already satisfied: scikit-learn in c:\users\penny\anaconda3\lib\site-packages (from Thermobar) (1.0.2)
Requirement already satisfied: scipy in c:\users\penny\anaconda3\lib\site-packages (from Thermobar) (1.9.1)
Requirement already satisfied: statsmodels in c:\users\penny\anaconda3\lib\site-packages (from Thermobar) (0.13.2)
Requirement already satisfied: openpyxl in c:\users\penny\anaconda3\lib\site-packages (from Thermobar) (3.0.10)
Requirement already satisfied: pyparsing>=2.2.1 in c:\users\penny\anaconda3\lib\site-packages (from matplotlib->Thermobar) (3.0.9)
Requirement already satisfied: packaging>=20.0 in c:\users\penny\anaconda3\lib\site-packages (from matplotlib->Thermobar) (21.3)
Requirement already satisfied: python-dateutil>=2.7 in c:\users\penny\anaconda3\lib\site-packages (from matplotlib->Thermobar) (2.8.2)
Requirement already satisfied: fonttools>=4.22.0 in c:\users\penny\anaconda3\lib\site-packages (from matplotlib->Thermobar) (4.25.0)
Requirement already satisfied: cycler>=0.10 in c:\users\penny\anaconda3\lib\site-packages (from matplotlib->Thermobar) (0.11.0)
Requirement already satisfied: pillow>=6.2.0 in c:\users\penny\anaconda3\lib\site-packages (from matplotlib->Thermobar) (9.2.0)
Requirement already satisfied: kiwisolver>=1.0.1 in c:\users\penny\anaconda3\lib\site-packages (from matplotlib->Thermobar) (1.4.2)
Requirement already satisfied: et_xmlfile in c:\users\penny\anaconda3\lib\site-packages (from openpyxl->Thermobar) (1.1.0)
Requirement already satisfied: pytz>=2020.1 in c:\users\penny\anaconda3\lib\site-packages (from pandas->Thermobar) (2022.1)
Requirement already satisfied: joblib>=0.11 in c:\users\penny\anaconda3\lib\site-packages (from scikit-learn->Thermobar) (1.1.0)
Requirement already satisfied: threadpoolctl>=2.0.0 in c:\users\penny\anaconda3\lib\site-packages (from scikit-learn->Thermobar) (2.2.0)
Requirement already satisfied: patsy>=0.5.2 in c:\users\penny\anaconda3\lib\site-packages (from statsmodels->Thermobar) (0.5.2)
Requirement already satisfied: six in c:\users\penny\anaconda3\lib\site-packages (from patsy>=0.5.2->statsmodels->Thermobar) (1.16.0)
This sets plotting parameters
[2]:
# This sets some plotting things
plt.rcParams["font.family"] = 'arial'
plt.rcParams["font.size"] =12
plt.rcParams["mathtext.default"] = "regular"
plt.rcParams["mathtext.fontset"] = "dejavusans"
plt.rcParams['patch.linewidth'] = 1
plt.rcParams['axes.linewidth'] = 1
plt.rcParams["xtick.direction"] = "in"
plt.rcParams["ytick.direction"] = "in"
plt.rcParams["ytick.direction"] = "in"
plt.rcParams["xtick.major.size"] = 6 # Sets length of ticks
plt.rcParams["ytick.major.size"] = 4 # Sets length of ticks
plt.rcParams["ytick.labelsize"] = 12 # Sets size of numbers on tick marks
plt.rcParams["xtick.labelsize"] = 12 # Sets size of numbers on tick marks
plt.rcParams["axes.titlesize"] = 14 # Overall title
plt.rcParams["axes.labelsize"] = 14 # Axes labels
Loading in Data
Gleeson et al (2020) had cpxs in the sheet “cpxs” and liquids in the “melts” tab
If you are loading your own data, your column headings need to be “Sample_ID”, “SiO2_Liq”, “MgO_Cpx” etc. e.g., oxide, then underscore, capital letter for phase. However, the order of columns doesnt matter
[3]:
# Loading Liquids
out=pt.import_excel('Gleeson2020JPET_Input_Pyroxene_Melts.xlsx', sheet_name="Melts")
my_input_Liqs=out['my_input']
myLiquids1=out['Liqs']
# Loading Cpxs from a different sheet
out2=pt.import_excel('Gleeson2020JPET_Input_Pyroxene_Melts.xlsx', sheet_name="Cpxs")
my_input_Cpxs=out2['my_input']
myCPXs1=out2['Cpxs']
# This loads in published barometry from Gleeson et al.
# You can delete this from your system, or swap it for something else useful.
Published=pd.read_excel('Gleeson2020JPET_Input_Pyroxene_Melts.xlsx', sheet_name="NP_Out")
[4]:
# You can check inputs have read in right using .head(). Check for columns of zeros where you expected data
display(myLiquids1.head())
display(myCPXs1.head())
| SiO2_Liq | TiO2_Liq | Al2O3_Liq | FeOt_Liq | MnO_Liq | MgO_Liq | CaO_Liq | Na2O_Liq | K2O_Liq | Cr2O3_Liq | P2O5_Liq | H2O_Liq | Fe3Fet_Liq | NiO_Liq | CoO_Liq | CO2_Liq | Sample_ID_Liq | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 47.1519 | 1.7168 | 15.5321 | 9.7208 | 0.1888 | 5.939515 | 12.3617 | 3.7556 | 1.1877 | 0.0 | 0.2766 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 17MMSG12_1 |
| 1 | 46.7277 | 1.7708 | 15.4931 | 9.5435 | 0.2096 | 6.098350 | 12.3699 | 3.7058 | 1.2644 | 0.0 | 0.1887 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 17MMSG12_2 |
| 2 | 47.5265 | 1.8483 | 15.7152 | 9.6930 | 0.1678 | 6.184563 | 12.3625 | 3.5107 | 1.2066 | 0.0 | 0.2055 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 17MMSG12_3 |
| 3 | 47.2916 | 1.7307 | 15.5250 | 9.3999 | 0.1588 | 6.322718 | 12.3696 | 3.9281 | 1.2285 | 0.0 | 0.2406 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 17MMSG12_4 |
| 4 | 47.2260 | 1.8009 | 15.6438 | 9.0440 | 0.2213 | 6.069612 | 12.4081 | 3.8352 | 1.1339 | 0.0 | 0.1894 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 17MMSG12_5 |
| SiO2_Cpx | TiO2_Cpx | Al2O3_Cpx | FeOt_Cpx | MnO_Cpx | MgO_Cpx | CaO_Cpx | Na2O_Cpx | K2O_Cpx | Cr2O3_Cpx | Sample_ID_Cpx | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 51.0169 | 0.5104 | 4.2921 | 3.9184 | 0.1135 | 15.9773 | 21.5003 | 0.3828 | 0.0 | 0.9642 | 17MMSG16_1 |
| 1 | 51.0208 | 0.5145 | 4.2768 | 3.7697 | 0.0917 | 15.9326 | 21.6712 | 0.3820 | 0.0 | 1.0514 | 17MMSG16_2 |
| 2 | 51.2990 | 0.4869 | 4.4177 | 3.7014 | 0.0983 | 15.9157 | 21.7450 | 0.3787 | 0.0 | 1.1905 | 17MMSG16_3 |
| 3 | 49.7147 | 0.7249 | 6.2489 | 3.9880 | 0.1128 | 15.0415 | 21.6397 | 0.4081 | 0.0 | 1.3550 | 17MMSG16_4 |
| 4 | 49.9807 | 0.7351 | 6.1948 | 4.0468 | 0.0951 | 14.9777 | 21.5061 | 0.3969 | 0.0 | 1.4862 | 17MMSG16_5 |
You can use help to get the documentation for this function to see the options/required inputs.
[5]:
help(pt.calculate_cpx_liq_press_temp_matching)
Help on function calculate_cpx_liq_press_temp_matching in module Thermobar.clinopyroxene_thermobarometry:
calculate_cpx_liq_press_temp_matching(*, liq_comps, cpx_comps, equationT=None, equationP=None, P=None, T=None, PMax=30, PMin=-10, Fe3Fet_Liq=None, Kd_Match='Putirka', Kd_Err=0.03, DiHd_Err=0.06, EnFs_Err=0.05, CaTs_Err=0.03, Cpx_Quality=False, H2O_Liq=None, return_all_pairs=False, iterations=30)
Evaluates all possible Opx-Liq pairs from N Liquids, M Cpx compositions
returns P (kbar) and T (K) for those in equilibrium.
Parameters
-----------
liq_comps: pandas.DataFrame
Panda DataFrame of liquid compositions with column headings SiO2_Liq etc.
cpx_comps: pandas.DataFrame
Panda DataFrame of cpx compositions with column headings SiO2_Cpx etc.
equationT: str
Specify equation for cpx thermometry (options):
| T_Put1996_eqT1 (P-indep, H2O-indep)
| T_Mas2013_eqTalk1 (P-indep, H2O-indep, alk adaption of T1)
| T_Brug2019 (P-indep, H2O-indep)
| T_Put1996_eqT2 (P-dep, H2O-indep)
| T_Mas2013_eqTalk2 (P-dep, H2O-indep, alk adaption of T2)
| T_Put1999 (P-dep, H2O-indep)
| T_Put2003 (P-dep, H2O-indep)
| T_Put1999 (P-dep, H2O-indep)
| T_Put2008_eq33 (P-dep, H2O-dep)
| T_Mas2013_eqalk33 (P-dep, H2O-dep, alk adaption of eq33)
| T_Mas2013_Palk2012 (P-indep, H2O_dep)
equationP: str
specify equation for cpx barometry (options):
| P_Put1996_eqP1 (T-dep, H2O-indep)
| P_Mas2013_eqPalk1 (T-dep, H2O-indep, alk adaption of P1)
| P_Put1996_eqP2 (T-dep, H2O-indep)
| P_Mas2013_eqPalk2 (T-dep, H2O-indep, alk adaption of P2)
| P_Put2003 ((T-dep, H2O-indep)
| P_Neave2017 (T-dep, H2O-indep)
| P_Put2008_eq30 (T-dep, H2O-dep)
| P_Put2008_eq31 (T-dep, H2O-dep)
| P_Put2008_eq32c (T-dep, H2O-dep)
| P_Mas2013_eqalk32c (T-dep, H2O-dep, alk adaption of 32c)
Or
T: int, float
Can also run calculations at a fixed temperature
P: int, float
Can also run calculations at a fixed pressure
Optional:
Kd_Match: int, str, optional
allows users to override the default of calculating Kd Fe-Mg based
on temperature using eq 35 of putirka
Set at fixed value (e.g., Kd_Match=0.27)
OR
specify Kd_Match=Masotta to use the Kd model fo Masotta et al. (2013),
which is also a function of Na and K, for trachytic and phonolitic magmas.
Kd_Err: int or float, Default=0.03
Allows users to specify the permitted error on Kd Fe-Mg (default=0.03 from Neave et al. 2019)
DiHd_Err: int or float, optional. Default=0.06
Allows users to specify the permitted error on DiHd (default=0.06 from Neave et al. 2019)
Compares measured and calculated values from Mollo et al. (2013)
EnFs_Err: int or float, optional. Default=0.05
Allows users to specify the permitted error on EnFs (default=0.05 from Neave et al. 2019)
Compares measured and calculated values from Mollo et al. (2013)
CaTs_Err: int or float, optional. Default=0.03
Allows users to specify the permitted error on CaTs (default=0.03 from Neave et al. 2019)
Compares measured and calculated values from Putirka (1999).
Fe3Fet_Liq: float, int, pandas.Series, optional
Fe3Fet ratio used to assess Kd Fe-Mg equilibrium between cpx and melt.
If users don't specify, uses Fe3Fet_Liq from liq_comps.
If specified, overwrites the Fe3Fet_Liq column in the liquid input.
H2O_Liq: float, int, pandas.Series, optional
If users don't specify, uses H2O_Liq from liq_comps, if specified overwrites this.
Cpx Quality: bool, optional
Default False. If True, filters out clinopyroxenes with cation sums outside of
4.02-3.99 (after Neave et al. 2017)
PMax: int or float, optional
Default value of 30 kbar. Uses to apply a preliminary KdFe-Mg filter
based on the T equation specified by the user.
PMin: int or float, optional
Default value of -10 kbar. Uses to apply a preliminary KdFe-Mg filter
based on the T equation specified by the user.
Returns: dict
Av_PTs: Average P and T for each cpx.
E.g., if cpx1 matches Liq1, Liq4, Liq6, Liq10, averages outputs for all 4 of those liquids.
Returns mean and 1 sigma of these averaged parameters for each Cpx.
All_PTs: Returns output parameters for all matches (e.g, cpx1-Liq1, cpx1-Liq4) without any averaging.
Example 1 - Default melt matching function options
Initially, we use the default, where all equilibrium criteria are considered.
We specify the H2O content of the liquid is 0.5 wt%
This returns a dictionary, containing 2 pandas dataframes. The first df labelled All_PTs gives the Pressures and temps for every single cpx-liq match, while the dataframe labelled Av_PTs averages all the parameters for each cpx. So if Cpx1 matches Liq1, Liq9, Liq 13, those three pressures and temperatures would be averaged.
[6]:
# Here we use Neave 2017 for P, and equation 33 from Putirka 2008 for T at H2O=0.5 wt%
MM1=pt.calculate_cpx_liq_press_temp_matching(liq_comps=myLiquids1, cpx_comps=myCPXs1,
equationP="P_Neave2017", equationT="T_Put2008_eq33",
H2O_Liq=0.5, iterations=30)
# These lines extract pandas dataframes from the dictionary MM1
# All matches
MM1_All=MM1['All_PTs']
# Averaged match per Cpx (following Neave...)
MM1_Av=MM1['Av_PTs']
Considering N=78 Cpx & N=163 Liqs, which is a total of N=12714 Liq-Cpx pairs, be patient if this is >>1 million!
3631 Matches remaining after initial Kd filter. Now moving onto iterative calculations
Finished calculating Ps and Ts, now just averaging the results. Almost there!
Done!!! I found a total of N=34 Cpx-Liq matches using the specified filter. N=3 Cpx out of the N=78 Cpx that you input matched to 1 or more liquids
Here, we inspect the average per Cpx, and see that only 3 Cpx analyses were found to have a match to any inputted liquid
One Cpx matches 28 liquids, another Cpx matches 4 liquids, and the final one matches 2 liquids.
You can tell from the Sample_ID_Cpx column which Cpxs these were (#64, 65, 66)
[11]:
display(MM1_Av)
| Sample_ID_Cpx | # of Liqs Averaged | Mean_T_K_calc | Std_T_K_calc | Mean_P_kbar_calc | Std_P_kbar_calc | ID_CPX | Mean_Delta_T_K_Iter | Mean_Delta_P_kbar_Iter | Mean_Delta_Kd_Put2008 | ... | Std_Delta_EnFs_I_M_Mollo13 | Std_CaTs_Pred_Put1999 | Std_Delta_CaTs_I_M_Put1999 | Std_CrCaTs_Pred_Put1999 | Std_Delta_CrCaTs_I_M_Put1999 | Std_CaTi_Pred_Put1999 | Std_Delta_CaTi_I_M_Put1999 | Std_Jd_Pred_Put1999 | Std_Delta_Jd_Put1999 | Std_Delta_Jd_I_M_Put1999 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 17MMSG16_64 | 28 | 1483.741762 | 20.458238 | 5.774846 | 0.598899 | 63 | 0.0 | 0.0 | 0.011725 | ... | 0.011676 | 0.000774 | 0.000774 | 0.0 | 0.0 | 0.003455 | 0.003334 | 0.001987 | 0.001987 | 0.001987 |
| 1 | 17MMSG16_65 | 4 | 1527.514642 | 7.965623 | 7.004365 | 0.163063 | 64 | 0.0 | 0.0 | 0.021093 | ... | 0.005158 | 0.000236 | 0.000236 | 0.0 | 0.0 | 0.002485 | 0.002485 | 0.000572 | 0.000572 | 0.000572 |
| 2 | 17MMSG16_66 | 2 | 1524.073696 | 6.637660 | 6.419271 | 0.146893 | 65 | 0.0 | 0.0 | 0.025916 | ... | 0.004697 | 0.000310 | 0.000310 | 0.0 | 0.0 | 0.000816 | 0.000816 | 0.000894 | 0.000894 | 0.000894 |
3 rows × 271 columns
We can also run calculations at a fixed temperature (1450 K)
[9]:
MM1_fixedT=pt.calculate_cpx_liq_press_temp_matching(liq_comps=myLiquids1, cpx_comps=myCPXs1,
equationP="P_Neave2017", T=1450,
H2O_Liq=0.5)
# These lines extract pandas dataframes from the dictionary MM1
MM1_All_fixedT=MM1_fixedT['All_PTs']
MM1_Av_fixedT=MM1_fixedT['Av_PTs']
Considering N=78 Cpx & N=163 Liqs, which is a total of N=12714 Liq-Cpx pairs, be patient if this is >>1 million!
Finished calculating Ps and Ts, now just averaging the results. Almost there!
Done!!! I found a total of N=42 Cpx-Liq matches using the specified filter. N=3 Cpx out of the N=78 Cpx that you input matched to 1 or more liquids
g:\my drive\postdoc\pymme\mybarometers\thermobar_outer\src\Thermobar\clinopyroxene_thermobarometry.py:2878: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
combo_liq_cpx_fur_filt['Delta_T_K_Iter']=Delta_T_K_Iter
g:\my drive\postdoc\pymme\mybarometers\thermobar_outer\src\Thermobar\clinopyroxene_thermobarometry.py:2879: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
combo_liq_cpx_fur_filt['Delta_P_kbar_Iter']=Delta_P_kbar_Iter
g:\my drive\postdoc\pymme\mybarometers\thermobar_outer\src\Thermobar\clinopyroxene_thermobarometry.py:2888: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
combo_liq_cpx_fur_filt.drop(["Sample_ID_Liq"], axis=1, inplace=True)
g:\my drive\postdoc\pymme\mybarometers\thermobar_outer\src\Thermobar\clinopyroxene_thermobarometry.py:2890: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
combo_liq_cpx_fur_filt.rename(columns={'T_K_calc': 'T_K_input'}, inplace=True)
[11]:
MM1_All_fixedT.head()
[11]:
| Sample_ID_Cpx | P_kbar_calc | T_K_input | Eq Tests Neave2017? | Delta_Kd_Put2008 | Delta_Kd_Mas2013 | Delta_EnFs_Mollo13 | Delta_EnFs_Put1999 | Delta_CaTs_Put1999 | Delta_DiHd_Mollo13 | ... | Delta_EnFs_I_M_Mollo13 | CaTs_Pred_Put1999 | Delta_CaTs_I_M_Put1999 | CrCaTs_Pred_Put1999 | Delta_CrCaTs_I_M_Put1999 | CaTi_Pred_Put1999 | Delta_CaTi_I_M_Put1999 | Jd_Pred_Put1999 | Delta_Jd_Put1999 | Delta_Jd_I_M_Put1999 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10317 | 17MMSG16_64 | 6.898724 | 1450 | True | 0.025861 | 0.079702 | 0.013569 | 0.004727 | 0.023827 | 0.046822 | ... | 0.013569 | 0.014860 | -0.023827 | 0.0 | 0.018583 | 0.034450 | 0.007844 | 0.008802 | 0.017738 | 0.017738 |
| 10324 | 17MMSG16_64 | 5.621206 | 1450 | True | 0.009732 | 0.109236 | 0.043536 | 0.016078 | 0.025944 | 0.016473 | ... | 0.043536 | 0.012743 | -0.025944 | 0.0 | 0.018583 | 0.028295 | 0.013999 | 0.010903 | 0.015637 | 0.015637 |
| 10328 | 17MMSG16_64 | 6.562306 | 1450 | True | 0.001378 | 0.070648 | 0.007186 | 0.006384 | 0.024758 | 0.015500 | ... | 0.007186 | 0.013928 | -0.024758 | 0.0 | 0.018583 | 0.030990 | 0.011304 | 0.008573 | 0.017967 | 0.017967 |
| 10329 | 17MMSG16_64 | 5.461770 | 1450 | True | 0.008311 | 0.095186 | 0.000852 | 0.002116 | 0.024891 | 0.000008 | ... | 0.000852 | 0.013796 | -0.024891 | 0.0 | 0.018583 | 0.034307 | 0.007987 | 0.011389 | 0.015151 | 0.015151 |
| 10330 | 17MMSG16_64 | 5.573106 | 1450 | True | 0.017770 | 0.109354 | 0.038498 | 0.017576 | 0.025572 | 0.011095 | ... | 0.038498 | 0.013115 | -0.025572 | 0.0 | 0.018583 | 0.025677 | 0.016617 | 0.010372 | 0.016167 | 0.016167 |
5 rows × 131 columns
Or at fixed pressure (5 kbar here)
[12]:
MM1_fixedP=pt.calculate_cpx_liq_press_temp_matching(liq_comps=myLiquids1, cpx_comps=myCPXs1,
P=5, equationT="T_Put2008_eq33",
H2O_Liq=0.5)
# These lines extract pandas dataframes from the dictionary MM1
MM1_All_fixedP=MM1_fixedP['All_PTs']
MM1_Av_fixedP=MM1_fixedP['Av_PTs']
Considering N=78 Cpx & N=163 Liqs, which is a total of N=12714 Liq-Cpx pairs, be patient if this is >>1 million!
Finished calculating Ps and Ts, now just averaging the results. Almost there!
Done!!! I found a total of N=34 Cpx-Liq matches using the specified filter. N=3 Cpx out of the N=78 Cpx that you input matched to 1 or more liquids
Example 1 b
We see that we only get 3 Cpx that matched. We might want to look at the distribution of equilibrium tests to work out what parameters to use
We can do this using return_all_pairs=True, then we can plot up the equilibrium test results
[13]:
# Here we use Neave 2017 for P, and equation 33 from Putirka 2008 for T at H2O=0.5 wt%
MM_allpairs=pt.calculate_cpx_liq_press_temp_matching(liq_comps=myLiquids1, cpx_comps=myCPXs1,
equationP="P_Neave2017", equationT="T_Put2008_eq33",
H2O_Liq=0.5, return_all_pairs=True)
# These lines extract pandas dataframes from the dictionary MM1
# All matches
MM_allpairs_All=MM_allpairs['All_PTs']
# Averaged match per Cpx (following Neave...)
MM_allpairs_Av=MM_allpairs['Av_PTs']
Considering N=78 Cpx & N=163 Liqs, which is a total of N=12714 Liq-Cpx pairs, be patient if this is >>1 million!
No equilibrium filters applied
Finished calculating Ps and Ts, now just averaging the results. Almost there!
Done!!! I found a total of N=12714 Cpx-Liq matches using the specified filter. N=78 Cpx out of the N=78 Cpx that you input matched to 1 or more liquids
[19]:
## You can see column names like this
MM_allpairs_Av.columns[0:100]
[19]:
Index(['Sample_ID_Cpx', '# of Liqs Averaged', 'Mean_T_K_calc', 'Std_T_K_calc',
'Mean_P_kbar_calc', 'Std_P_kbar_calc', 'ID_CPX',
'Mean_Delta_Kd_Put2008', 'Mean_Delta_Kd_Mas2013',
'Mean_Delta_EnFs_Mollo13', 'Mean_Delta_EnFs_Put1999',
'Mean_Delta_CaTs_Put1999', 'Mean_Delta_DiHd_Mollo13',
'Mean_Delta_DiHd_Put1999', 'Mean_Delta_CrCaTs_Put1999',
'Mean_Delta_CaTi_Put1999', 'Mean_SiO2_Liq', 'Mean_TiO2_Liq',
'Mean_Al2O3_Liq', 'Mean_FeOt_Liq', 'Mean_MnO_Liq', 'Mean_MgO_Liq',
'Mean_CaO_Liq', 'Mean_Na2O_Liq', 'Mean_K2O_Liq', 'Mean_Cr2O3_Liq',
'Mean_P2O5_Liq', 'Mean_H2O_Liq', 'Mean_Fe3Fet_Liq', 'Mean_NiO_Liq',
'Mean_CoO_Liq', 'Mean_CO2_Liq', 'Mean_SiO2_Liq_mol_frac',
'Mean_MgO_Liq_mol_frac', 'Mean_MnO_Liq_mol_frac',
'Mean_FeOt_Liq_mol_frac', 'Mean_CaO_Liq_mol_frac',
'Mean_Al2O3_Liq_mol_frac', 'Mean_Na2O_Liq_mol_frac',
'Mean_K2O_Liq_mol_frac', 'Mean_TiO2_Liq_mol_frac',
'Mean_P2O5_Liq_mol_frac', 'Mean_Cr2O3_Liq_mol_frac',
'Mean_Si_Liq_cat_frac', 'Mean_Mg_Liq_cat_frac', 'Mean_Mn_Liq_cat_frac',
'Mean_Fet_Liq_cat_frac', 'Mean_Ca_Liq_cat_frac', 'Mean_Al_Liq_cat_frac',
'Mean_Na_Liq_cat_frac', 'Mean_K_Liq_cat_frac', 'Mean_Ti_Liq_cat_frac',
'Mean_P_Liq_cat_frac', 'Mean_Cr_Liq_cat_frac',
'Mean_Mg_Number_Liq_NoFe3', 'Mean_Mg_Number_Liq_Fe3', 'Mean_ID_Liq',
'Mean_SiO2_Cpx', 'Mean_TiO2_Cpx', 'Mean_Al2O3_Cpx', 'Mean_FeOt_Cpx',
'Mean_MnO_Cpx', 'Mean_MgO_Cpx', 'Mean_CaO_Cpx', 'Mean_Na2O_Cpx',
'Mean_K2O_Cpx', 'Mean_Cr2O3_Cpx', 'Mean_Si_Cpx_cat_6ox',
'Mean_Mg_Cpx_cat_6ox', 'Mean_Fet_Cpx_cat_6ox', 'Mean_Ca_Cpx_cat_6ox',
'Mean_Al_Cpx_cat_6ox', 'Mean_Na_Cpx_cat_6ox', 'Mean_K_Cpx_cat_6ox',
'Mean_Mn_Cpx_cat_6ox', 'Mean_Ti_Cpx_cat_6ox', 'Mean_Cr_Cpx_cat_6ox',
'Mean_oxy_renorm_factor', 'Mean_Al_IV_cat_6ox', 'Mean_Al_VI_cat_6ox',
'Mean_En_Simple_MgFeCa_Cpx', 'Mean_Fs_Simple_MgFeCa_Cpx',
'Mean_Wo_Simple_MgFeCa_Cpx', 'Mean_Cation_Sum_Cpx', 'Mean_Ca_CaMgFe',
'Mean_Lindley_Fe3_Cpx', 'Mean_Lindley_Fe2_Cpx',
'Mean_Lindley_Fe3_Cpx_prop', 'Mean_CrCaTs', 'Mean_a_cpx_En',
'Mean_Mgno_Cpx', 'Mean_Jd', 'Mean_Jd_from 0=Na, 1=Al', 'Mean_CaTs',
'Mean_CaTi', 'Mean_DiHd_1996', 'Mean_EnFs', 'Mean_DiHd_2003',
'Mean_Di_Cpx', 'Mean_FeIII_Wang21'],
dtype='object')
[22]:
fig, ((ax1, ax2), (ax3, ax4)) = plt.subplots(2,2, figsize = (12,10))
ax1.hist(MM_allpairs_All['Delta_Kd_Put2008'], ec='k')
ax1.set_xlabel('Delta KD Putirka (2008)')
ax2.hist(MM_allpairs_All['Delta_EnFs_Mollo13'], ec='k')
ax2.set_xlabel('Delta_EnFs_Mollo13')
ax3.hist(MM_allpairs_All['Delta_DiHd_Mollo13'], ec='k')
ax3.set_xlabel('Delta_DiHd_Mollo13')
ax4.hist(MM_allpairs_All['Delta_CaTs_Put1999'], ec='k')
ax4.set_xlabel('Delta_CaTs_Put1999')
[22]:
Text(0.5, 0, 'Delta_CaTs_Put1999')
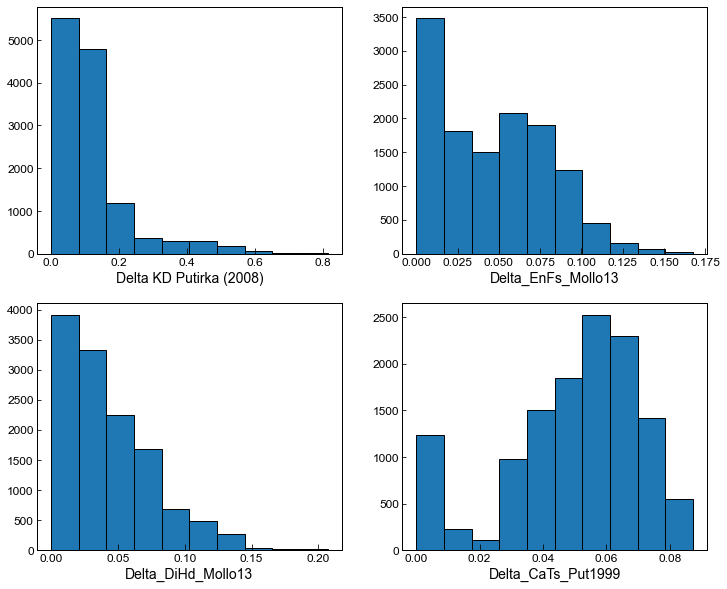
We can adapt the filters to get more matches
Here, folowing Gleeson et al. 2020, we specify that we wish to consider pairs which pass the DiHd, CaTs and EnFs equilibrium tests with sigma = 2.
To make the function most customizable, instead of just entering a sigma, you can enter an _Err for each equilibrium test to define what filters you want. E.g. here we consider matches within +/- 0.03 for Kd, but +-0.06 for CaTs, DiHd, and +-0.1 for EnFs. These are the “pass” values from Neave et al. (2019) for CaTs, DiHd and EnFs multiplied by 2
[23]:
MM1_2s=pt.calculate_cpx_liq_press_temp_matching(liq_comps=myLiquids1, cpx_comps=myCPXs1,
equationP="P_Neave2017", equationT="T_Put2008_eq33",
Kd_Err=0.03, EnFs_Err=0.1, DiHd_Err=0.12,
CaTs_Err=0.06,
H2O_Liq=0.5)
# These lines extract pandas dataframes from the dictionary MM1
MM1_2s_All=MM1_2s['All_PTs']
MM1_2s_Av=MM1_2s['Av_PTs']
Considering N=78 Cpx & N=163 Liqs, which is a total of N=12714 Liq-Cpx pairs, be patient if this is >>1 million!
3892 Matches remaining after initial Kd filter. Now moving onto iterative calculations
Finished calculating Ps and Ts, now just averaging the results. Almost there!
Done!!! I found a total of N=1008 Cpx-Liq matches using the specified filter. N=54 Cpx out of the N=78 Cpx that you input matched to 1 or more liquids
We can look and see we get a lot more matches.
[24]:
MM1_2s_Av
[24]:
| Sample_ID_Cpx | # of Liqs Averaged | Mean_T_K_calc | Std_T_K_calc | Mean_P_kbar_calc | Std_P_kbar_calc | ID_CPX | Mean_Delta_Kd_Put2008 | Mean_Delta_Kd_Mas2013 | Mean_Delta_EnFs_Mollo13 | ... | Std_Delta_EnFs_I_M_Mollo13 | Std_CaTs_Pred_Put1999 | Std_Delta_CaTs_I_M_Put1999 | Std_CrCaTs_Pred_Put1999 | Std_Delta_CrCaTs_I_M_Put1999 | Std_CaTi_Pred_Put1999 | Std_Delta_CaTi_I_M_Put1999 | Std_Jd_Pred_Put1999 | Std_Delta_Jd_Put1999 | Std_Delta_Jd_I_M_Put1999 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 17MMSG16_1 | 41 | 1485.889871 | 24.563876 | 6.181435 | 0.702366 | 0 | 0.017003 | 0.109085 | 0.034090 | ... | 0.015727 | 0.001063 | 0.001063 | 0.0 | 0.0 | 0.003347 | 0.002276 | 0.001916 | 0.001916 | 0.001916 |
| 1 | 17MMSG16_2 | 42 | 1481.111278 | 22.899738 | 6.043041 | 0.675803 | 1 | 0.014476 | 0.108837 | 0.040860 | ... | 0.016491 | 0.001058 | 0.001058 | 0.0 | 0.0 | 0.003308 | 0.002076 | 0.001858 | 0.001858 | 0.001858 |
| 2 | 17MMSG16_3 | 48 | 1478.881332 | 20.342782 | 5.946874 | 0.672108 | 2 | 0.014329 | 0.107455 | 0.048397 | ... | 0.016401 | 0.001067 | 0.001067 | 0.0 | 0.0 | 0.003941 | 0.002634 | 0.001926 | 0.001926 | 0.001926 |
| 3 | 17MMSG16_4 | 12 | 1508.719399 | 13.984144 | 7.269439 | 0.698513 | 3 | 0.016959 | 0.113074 | 0.041563 | ... | 0.010652 | 0.001745 | 0.001745 | 0.0 | 0.0 | 0.002842 | 0.002842 | 0.001712 | 0.001712 | 0.001712 |
| 4 | 17MMSG16_5 | 2 | 1504.328416 | 12.762583 | 7.987680 | 0.609534 | 4 | 0.015726 | 0.112612 | 0.035519 | ... | 0.017390 | 0.002786 | 0.002786 | 0.0 | 0.0 | 0.004185 | 0.004185 | 0.001962 | 0.001962 | 0.001962 |
| 5 | 17MMSG16_6 | 11 | 1468.442918 | 27.311896 | 6.360127 | 0.481471 | 5 | 0.017424 | 0.112557 | 0.060445 | ... | 0.023139 | 0.001463 | 0.001463 | 0.0 | 0.0 | 0.009994 | 0.005090 | 0.002061 | 0.002061 | 0.002061 |
| 6 | 17MMSG16_7 | 1 | 1488.553479 | NaN | 7.293616 | NaN | 6 | 0.017285 | 0.106398 | 0.026222 | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 7 | 17MMSG16_8 | 3 | 1497.688206 | 24.208102 | 7.131719 | 0.458597 | 7 | 0.022747 | 0.106427 | 0.032062 | ... | 0.013816 | 0.000806 | 0.000806 | 0.0 | 0.0 | 0.017912 | 0.001510 | 0.004366 | 0.004366 | 0.004366 |
| 8 | 17MMSG16_9 | 14 | 1478.123890 | 30.729723 | 7.116592 | 0.838216 | 8 | 0.013937 | 0.111672 | 0.057300 | ... | 0.019389 | 0.002407 | 0.002407 | 0.0 | 0.0 | 0.003769 | 0.003769 | 0.002502 | 0.002502 | 0.002502 |
| 9 | 17MMSG16_15 | 41 | 1488.886000 | 26.181968 | 6.703865 | 0.716215 | 14 | 0.018076 | 0.108948 | 0.045087 | ... | 0.016411 | 0.001136 | 0.001136 | 0.0 | 0.0 | 0.003261 | 0.002201 | 0.001912 | 0.001912 | 0.001912 |
| 10 | 17MMSG16_16 | 42 | 1487.353992 | 23.120144 | 6.116487 | 0.680899 | 15 | 0.014177 | 0.107697 | 0.033822 | ... | 0.017744 | 0.001130 | 0.001130 | 0.0 | 0.0 | 0.003194 | 0.002012 | 0.001858 | 0.001858 | 0.001858 |
| 11 | 17MMSG16_17 | 38 | 1488.119090 | 26.755005 | 6.347794 | 0.710585 | 16 | 0.018082 | 0.110828 | 0.045613 | ... | 0.017549 | 0.001190 | 0.001190 | 0.0 | 0.0 | 0.003193 | 0.002312 | 0.001866 | 0.001866 | 0.001866 |
| 12 | 17MMSG16_18 | 24 | 1486.626170 | 30.720472 | 6.596753 | 0.797696 | 17 | 0.017821 | 0.107680 | 0.047007 | ... | 0.019286 | 0.001817 | 0.001817 | 0.0 | 0.0 | 0.003515 | 0.002275 | 0.002127 | 0.002127 | 0.002127 |
| 13 | 17MMSG16_19 | 31 | 1488.144727 | 29.126235 | 6.596542 | 0.690676 | 18 | 0.017452 | 0.107400 | 0.035286 | ... | 0.017125 | 0.001087 | 0.001087 | 0.0 | 0.0 | 0.003583 | 0.002398 | 0.001913 | 0.001913 | 0.001913 |
| 14 | 17MMSG16_23 | 2 | 1516.099263 | 8.783466 | 11.129027 | 0.922784 | 22 | 0.015429 | 0.024621 | 0.007368 | ... | 0.000384 | 0.000435 | 0.000435 | 0.0 | 0.0 | 0.003083 | 0.003083 | 0.001316 | 0.001316 | 0.001316 |
| 15 | 17MMSG16_25 | 17 | 1487.199233 | 30.379454 | 7.797178 | 0.804796 | 24 | 0.014354 | 0.110045 | 0.040049 | ... | 0.017164 | 0.002152 | 0.002152 | 0.0 | 0.0 | 0.003870 | 0.003714 | 0.002319 | 0.002319 | 0.002319 |
| 16 | 17MMSG16_26 | 2 | 1517.033574 | 13.157843 | 9.192633 | 0.638309 | 25 | 0.016485 | 0.122640 | 0.024599 | ... | 0.016140 | 0.002642 | 0.002642 | 0.0 | 0.0 | 0.004291 | 0.000696 | 0.001962 | 0.001962 | 0.001962 |
| 17 | 17MMSG16_27 | 13 | 1468.680275 | 32.487084 | 6.201820 | 0.807941 | 26 | 0.016732 | 0.110118 | 0.055827 | ... | 0.019313 | 0.002292 | 0.002292 | 0.0 | 0.0 | 0.003975 | 0.003975 | 0.002578 | 0.002578 | 0.002578 |
| 18 | 17MMSG16_28 | 1 | 1481.335296 | NaN | 6.628711 | NaN | 27 | 0.012402 | 0.111280 | 0.024218 | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 19 | 17MMSG16_31 | 4 | 1484.167312 | 22.260242 | 8.608298 | 1.158593 | 30 | 0.018221 | 0.077493 | 0.008053 | ... | 0.006173 | 0.001667 | 0.001667 | 0.0 | 0.0 | 0.011392 | 0.010853 | 0.003890 | 0.003890 | 0.003890 |
| 20 | 17MMSG16_32 | 3 | 1493.347508 | 15.766560 | 9.022041 | 1.021551 | 31 | 0.020157 | 0.056568 | 0.009968 | ... | 0.005500 | 0.001333 | 0.001333 | 0.0 | 0.0 | 0.005092 | 0.005092 | 0.002447 | 0.002447 | 0.002447 |
| 21 | 17MMSG16_33 | 7 | 1466.794930 | 26.303530 | 7.771733 | 1.082330 | 32 | 0.022224 | 0.076776 | 0.011582 | ... | 0.019652 | 0.003312 | 0.003312 | 0.0 | 0.0 | 0.012156 | 0.012156 | 0.005247 | 0.005247 | 0.005247 |
| 22 | 17MMSG16_41 | 2 | 1484.939273 | 12.232729 | 6.729527 | 0.575435 | 40 | 0.015522 | 0.112878 | 0.028812 | ... | 0.015094 | 0.002203 | 0.002203 | 0.0 | 0.0 | 0.004677 | 0.003916 | 0.001961 | 0.001961 | 0.001961 |
| 23 | 17MMSG16_42 | 15 | 1470.419150 | 30.552638 | 6.507811 | 0.799564 | 41 | 0.013192 | 0.109001 | 0.034984 | ... | 0.016686 | 0.002044 | 0.002044 | 0.0 | 0.0 | 0.004277 | 0.004010 | 0.002403 | 0.002403 | 0.002403 |
| 24 | 17MMSG16_43 | 3 | 1487.005177 | 22.572010 | 7.104371 | 0.872211 | 42 | 0.020541 | 0.105120 | 0.013477 | ... | 0.012230 | 0.002087 | 0.002087 | 0.0 | 0.0 | 0.017480 | 0.015351 | 0.005004 | 0.005004 | 0.005004 |
| 25 | 17MMSG16_44 | 14 | 1466.049303 | 6.756044 | 5.164047 | 0.138327 | 43 | 0.020988 | 0.121122 | 0.060390 | ... | 0.002272 | 0.000392 | 0.000392 | 0.0 | 0.0 | 0.005397 | 0.005397 | 0.000948 | 0.000948 | 0.000948 |
| 26 | 17MMSG16_46 | 1 | 1450.992064 | NaN | 6.131601 | NaN | 45 | 0.022115 | 0.126785 | 0.019921 | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 27 | 17MMSG16_47 | 6 | 1478.389492 | 30.427140 | 7.919723 | 1.160778 | 46 | 0.018992 | 0.094339 | 0.012751 | ... | 0.019967 | 0.003332 | 0.003332 | 0.0 | 0.0 | 0.009684 | 0.009684 | 0.003889 | 0.003889 | 0.003889 |
| 28 | 17MMSG16_48 | 6 | 1461.641359 | 25.435532 | 6.906666 | 1.075573 | 47 | 0.017849 | 0.087092 | 0.013369 | ... | 0.018997 | 0.002753 | 0.002753 | 0.0 | 0.0 | 0.009734 | 0.009734 | 0.003593 | 0.003593 | 0.003593 |
| 29 | 17MMSG16_49 | 4 | 1459.812687 | 21.434169 | 7.278039 | 0.511793 | 48 | 0.019831 | 0.102896 | 0.014924 | ... | 0.021996 | 0.002708 | 0.002708 | 0.0 | 0.0 | 0.012413 | 0.007813 | 0.002384 | 0.002384 | 0.002384 |
| 30 | 17MMSG16_50 | 5 | 1465.445525 | 19.200524 | 7.625287 | 0.449292 | 49 | 0.017306 | 0.096452 | 0.013228 | ... | 0.019041 | 0.002445 | 0.002445 | 0.0 | 0.0 | 0.010740 | 0.008390 | 0.002292 | 0.002292 | 0.002292 |
| 31 | 17MMSG16_51 | 5 | 1469.018640 | 28.869263 | 7.788495 | 1.207809 | 50 | 0.019463 | 0.089174 | 0.014356 | ... | 0.021085 | 0.003127 | 0.003127 | 0.0 | 0.0 | 0.010777 | 0.010777 | 0.004006 | 0.004006 | 0.004006 |
| 32 | 17MMSG16_52 | 6 | 1470.735693 | 25.889162 | 7.709866 | 1.098808 | 51 | 0.018743 | 0.084041 | 0.013599 | ... | 0.019372 | 0.002875 | 0.002875 | 0.0 | 0.0 | 0.009529 | 0.009415 | 0.003594 | 0.003594 | 0.003594 |
| 33 | 17MMSG16_53 | 4 | 1461.603342 | 21.507755 | 7.497221 | 0.515600 | 52 | 0.019794 | 0.101870 | 0.016555 | ... | 0.022205 | 0.002752 | 0.002752 | 0.0 | 0.0 | 0.012317 | 0.008822 | 0.002384 | 0.002384 | 0.002384 |
| 34 | 17MMSG16_54 | 6 | 1468.339060 | 25.775449 | 7.560598 | 1.093696 | 53 | 0.018288 | 0.085621 | 0.014316 | ... | 0.019271 | 0.002847 | 0.002847 | 0.0 | 0.0 | 0.009574 | 0.005766 | 0.003594 | 0.003594 | 0.003594 |
| 35 | 17MMSG16_55 | 6 | 1475.419684 | 26.119400 | 8.068702 | 1.109918 | 54 | 0.017422 | 0.088739 | 0.013183 | ... | 0.019618 | 0.002941 | 0.002941 | 0.0 | 0.0 | 0.009423 | 0.008640 | 0.003594 | 0.003594 | 0.003594 |
| 36 | 17MMSG16_56 | 6 | 1467.958418 | 26.131909 | 7.400169 | 1.104021 | 55 | 0.020948 | 0.080256 | 0.012871 | ... | 0.020907 | 0.002898 | 0.002898 | 0.0 | 0.0 | 0.009950 | 0.008418 | 0.003620 | 0.003620 | 0.003620 |
| 37 | 17MMSG16_57 | 5 | 1477.542978 | 19.061539 | 8.235879 | 1.005935 | 56 | 0.018641 | 0.071813 | 0.010880 | ... | 0.006270 | 0.001389 | 0.001389 | 0.0 | 0.0 | 0.010346 | 0.009841 | 0.003371 | 0.003371 | 0.003371 |
| 38 | 17MMSG16_58 | 5 | 1477.558853 | 19.608754 | 8.134983 | 1.054776 | 57 | 0.017729 | 0.078405 | 0.011768 | ... | 0.006724 | 0.001446 | 0.001446 | 0.0 | 0.0 | 0.010083 | 0.009600 | 0.003371 | 0.003371 | 0.003371 |
| 39 | 17MMSG16_60 | 1 | 1510.282641 | NaN | 9.941902 | NaN | 59 | 0.024036 | 0.027654 | 0.005248 | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 40 | 17MMSG16_63 | 14 | 1500.519635 | 15.260532 | 6.416086 | 0.696749 | 62 | 0.015229 | 0.102342 | 0.048159 | ... | 0.010220 | 0.000660 | 0.000660 | 0.0 | 0.0 | 0.003081 | 0.003081 | 0.001665 | 0.001665 | 0.001665 |
| 41 | 17MMSG16_64 | 51 | 1477.815669 | 22.546369 | 5.778123 | 0.650738 | 63 | 0.014993 | 0.101857 | 0.045509 | ... | 0.017546 | 0.001118 | 0.001118 | 0.0 | 0.0 | 0.003737 | 0.003667 | 0.001768 | 0.001768 | 0.001768 |
| 42 | 17MMSG16_65 | 59 | 1484.092363 | 20.177984 | 6.336113 | 0.609397 | 64 | 0.014676 | 0.101845 | 0.051491 | ... | 0.017277 | 0.001121 | 0.001121 | 0.0 | 0.0 | 0.003646 | 0.003220 | 0.001727 | 0.001727 | 0.001727 |
| 43 | 17MMSG16_66 | 59 | 1480.284084 | 20.014002 | 5.790472 | 0.601774 | 65 | 0.014602 | 0.102233 | 0.043788 | ... | 0.017151 | 0.001095 | 0.001095 | 0.0 | 0.0 | 0.003690 | 0.002957 | 0.001727 | 0.001727 | 0.001727 |
| 44 | 17MMSG16_67 | 53 | 1481.980241 | 20.231170 | 5.970434 | 0.636838 | 66 | 0.015777 | 0.101115 | 0.044789 | ... | 0.016580 | 0.001104 | 0.001104 | 0.0 | 0.0 | 0.003672 | 0.003516 | 0.001691 | 0.001691 | 0.001691 |
| 45 | 17MMSG16_68 | 47 | 1481.370873 | 21.662019 | 6.333638 | 0.672169 | 67 | 0.017513 | 0.110048 | 0.046434 | ... | 0.014251 | 0.000988 | 0.000988 | 0.0 | 0.0 | 0.003448 | 0.003122 | 0.001912 | 0.001912 | 0.001912 |
| 46 | 17MMSG16_69 | 17 | 1473.621605 | 33.602175 | 6.452342 | 0.842672 | 68 | 0.014151 | 0.105720 | 0.052688 | ... | 0.018308 | 0.002037 | 0.002037 | 0.0 | 0.0 | 0.004104 | 0.004104 | 0.002563 | 0.002563 | 0.002563 |
| 47 | 17MMSG16_70 | 38 | 1482.932831 | 26.888222 | 6.325916 | 0.729869 | 69 | 0.017481 | 0.109445 | 0.038525 | ... | 0.015194 | 0.001061 | 0.001061 | 0.0 | 0.0 | 0.003433 | 0.002620 | 0.001956 | 0.001956 | 0.001956 |
| 48 | 17MMSG16_71 | 36 | 1480.126635 | 28.341597 | 6.112875 | 0.697415 | 70 | 0.018407 | 0.110369 | 0.044493 | ... | 0.015750 | 0.001049 | 0.001049 | 0.0 | 0.0 | 0.003518 | 0.002461 | 0.001872 | 0.001872 | 0.001872 |
| 49 | 17MMSG16_72 | 49 | 1477.049409 | 23.385781 | 5.736961 | 0.716733 | 71 | 0.014679 | 0.106935 | 0.045030 | ... | 0.018130 | 0.001135 | 0.001135 | 0.0 | 0.0 | 0.004040 | 0.002541 | 0.002031 | 0.002031 | 0.002031 |
| 50 | 17MMSG16_73 | 21 | 1465.895725 | 32.044502 | 5.619340 | 0.782025 | 72 | 0.014084 | 0.107360 | 0.040519 | ... | 0.016108 | 0.001692 | 0.001692 | 0.0 | 0.0 | 0.003906 | 0.002626 | 0.002380 | 0.002380 | 0.002380 |
| 51 | 17MMSG16_74 | 27 | 1471.586727 | 29.272904 | 5.505243 | 0.697361 | 73 | 0.016905 | 0.106057 | 0.038048 | ... | 0.016442 | 0.001438 | 0.001438 | 0.0 | 0.0 | 0.003920 | 0.002876 | 0.001977 | 0.001977 | 0.001977 |
| 52 | 17MMSG16_75 | 36 | 1478.943758 | 28.315156 | 6.233307 | 0.699542 | 74 | 0.018136 | 0.109754 | 0.041010 | ... | 0.014589 | 0.000991 | 0.000991 | 0.0 | 0.0 | 0.003641 | 0.002419 | 0.001872 | 0.001872 | 0.001872 |
| 53 | 17MMSG16_78 | 2 | 1447.236817 | 2.337629 | 5.104677 | 0.108849 | 77 | 0.017112 | 0.121629 | 0.041069 | ... | 0.001189 | 0.000188 | 0.000188 | 0.0 | 0.0 | 0.010952 | 0.010952 | 0.000930 | 0.000930 | 0.000930 |
54 rows × 267 columns
Example 2 - Fixed Kd value, rather than function of Temp
Say you want to used a fixed value for Kd Fe-Mg, rather than the default, which uses equation 35 of Putirka which is T dependent
Here, specifying Kd Fe-Mg = 0.27, and you want to consider +-0.08 as an equilibrium match
Also specifying here that Fe3Fet_Liq=0.15
Also changing pressure equation to equation 30
Using 2 sigma values from Neave et al. (2019) for DiHd, CaTs, EnFs
[25]:
MM2=pt.calculate_cpx_liq_press_temp_matching(liq_comps=myLiquids1, cpx_comps=myCPXs1,
equationP="P_Put2008_eq30", equationT="T_Put2008_eq33",
Kd_Match=0.27, Kd_Err=0.08, Fe3Fet_Liq=0.15, CaTs_Err=0.06, DiHd_Err=0.12, EnFs_Err=0.06)
MM2_All=MM2['All_PTs']
MM2_Av=MM2['Av_PTs']
Considering N=78 Cpx & N=163 Liqs, which is a total of N=12714 Liq-Cpx pairs, be patient if this is >>1 million!
the code is evaluating Kd matches using Kd=0.27
4172 Matches remaining after initial Kd filter. Now moving onto iterative calculations
Finished calculating Ps and Ts, now just averaging the results. Almost there!
Done!!! I found a total of N=2554 Cpx-Liq matches using the specified filter. N=57 Cpx out of the N=78 Cpx that you input matched to 1 or more liquids
Example 3 - Alkaline Systems
You might want to use the updated Kd model from Masotta calibrated for trachyte and phonolitic magmas.
Here, you can change Kd_Match to “Masotta”
Might want to use P from P_Mas2013_Palk2012 and T_Mas2013_Talk2012
[26]:
MM3=pt.calculate_cpx_liq_press_temp_matching(liq_comps=myLiquids1, cpx_comps=myCPXs1,
equationP="P_Mas2013_Palk2012", equationT="T_Mas2013_Talk2012",
Kd_Match="Masotta", Kd_Err=0.08, Fe3Fet_Liq=0.15, CaTs_Err=0.06, DiHd_Err=0.12, EnFs_Err=0.06)
MM3_All=MM3['All_PTs']
MM3_Av=MM3['Av_PTs']
Caution, you have selected to use the Kd-Fe-Mg model of Masotta et al. (2013)which is only valid for trachyte and phonolitic magmas. use PutKd=True to use the Kd model of Putirka (2008)
Considering N=78 Cpx & N=163 Liqs, which is a total of N=12714 Liq-Cpx pairs, be patient if this is >>1 million!
Youve selected a P-independent function
Youve selected a P-independent function
2352 Matches remaining after initial Kd filter. Now moving onto iterative calculations
Youve selected a P-independent function
Youve selected a T-independent function
Youve selected a T-independent function
Finished calculating Ps and Ts, now just averaging the results. Almost there!
Done!!! I found a total of N=1580 Cpx-Liq matches using the specified filter. N=50 Cpx out of the N=78 Cpx that you input matched to 1 or more liquids
[27]:
MM3_All.head()
[27]:
| Sample_ID_Cpx | P_kbar_calc | T_K_calc | Eq Tests Neave2017? | Delta_Kd_Put2008 | Delta_Kd_Mas2013 | Delta_EnFs_Mollo13 | Delta_EnFs_Put1999 | Delta_CaTs_Put1999 | Delta_DiHd_Mollo13 | ... | Delta_EnFs_I_M_Mollo13 | CaTs_Pred_Put1999 | Delta_CaTs_I_M_Put1999 | CrCaTs_Pred_Put1999 | Delta_CrCaTs_I_M_Put1999 | CaTi_Pred_Put1999 | Delta_CaTi_I_M_Put1999 | Jd_Pred_Put1999 | Delta_Jd_Put1999 | Delta_Jd_I_M_Put1999 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 17MMSG16_1 | 4.484954 | 1353.763834 | False | 0.076093 | 0.027189 | 0.028005 | 0.005255 | 0.033390 | 0.023960 | ... | -0.028005 | 0.014322 | -0.033390 | 0.0 | 0.014105 | 0.065141 | 0.032986 | 0.017433 | 0.010033 | 0.010033 |
| 1 | 17MMSG16_1 | 4.536992 | 1359.812658 | False | 0.069446 | 0.030948 | 0.027431 | 0.007617 | 0.033259 | 0.018175 | ... | -0.027431 | 0.014452 | -0.033259 | 0.0 | 0.014105 | 0.066164 | 0.034008 | 0.017281 | 0.010185 | 0.010185 |
| 2 | 17MMSG16_1 | 4.664336 | 1362.026843 | False | 0.070245 | 0.029827 | 0.024555 | 0.004171 | 0.033089 | 0.012565 | ... | -0.024555 | 0.014623 | -0.033089 | 0.0 | 0.014105 | 0.063032 | 0.030877 | 0.016213 | 0.011253 | 0.011253 |
| 3 | 17MMSG16_1 | 4.521748 | 1361.019784 | False | 0.060027 | 0.043562 | 0.025938 | 0.007389 | 0.033548 | 0.021885 | ... | -0.025938 | 0.014163 | -0.033548 | 0.0 | 0.014105 | 0.065681 | 0.033525 | 0.018096 | 0.009371 | 0.009371 |
| 4 | 17MMSG16_1 | 4.523901 | 1356.789776 | False | 0.059466 | 0.046108 | 0.028110 | 0.008869 | 0.033104 | 0.018335 | ... | -0.028110 | 0.014608 | -0.033104 | 0.0 | 0.014105 | 0.069209 | 0.037054 | 0.017837 | 0.009630 | 0.009630 |
5 rows × 133 columns
Example 4 - Say you only want a Kd filter, using the equation of Putirka
Kd_Match=“Putirka” is actually a default option, so you don’t have to type it
Easiest way to do this without having a lot of effort in the function is just to set very high “pass” values for the other filters.
[28]:
MM4=pt.calculate_cpx_liq_press_temp_matching(liq_comps=myLiquids1, cpx_comps=myCPXs1,
equationP="P_Neave2017", equationT="T_Put2008_eq33",
Kd_Match="Putirka", Kd_Err=0.03, Fe3Fet_Liq=0.15,
CaTs_Err=10, DiHd_Err=10, EnFs_Err=10)
MM4_All=MM4['All_PTs']
MM4_Av=MM4['Av_PTs']
Considering N=78 Cpx & N=163 Liqs, which is a total of N=12714 Liq-Cpx pairs, be patient if this is >>1 million!
3043 Matches remaining after initial Kd filter. Now moving onto iterative calculations
Finished calculating Ps and Ts, now just averaging the results. Almost there!
Done!!! I found a total of N=1265 Cpx-Liq matches using the specified filter. N=69 Cpx out of the N=78 Cpx that you input matched to 1 or more liquids
Example 5 - Plotting Equilibrium filters
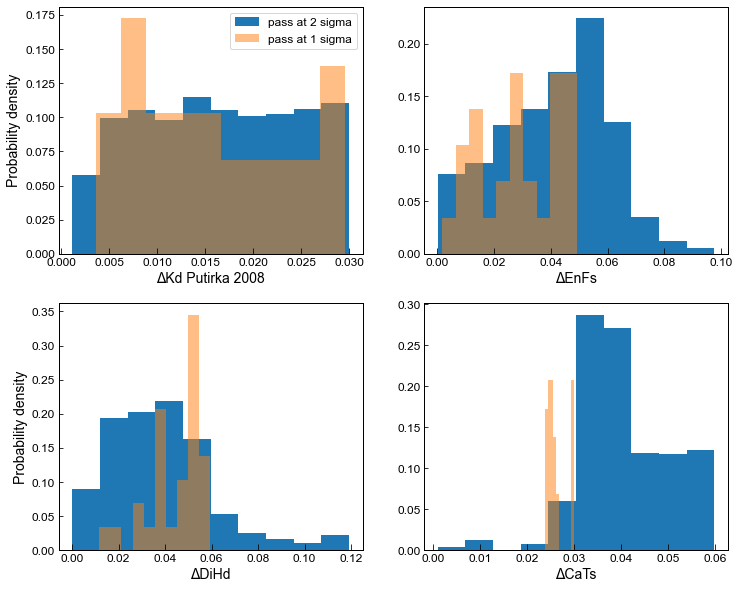
Here, we compare results from the default filter (stored in MM1_All) and those using 2* preferred values (stored in MM1_2s_All)
[29]:
fig, ((ax1, ax2), (ax3, ax4)) = plt.subplots(2, 2, figsize=(12, 10))
# This plots distribution of Kd in matches with probability on the y axis
ax1.hist(MM1_2s_All['Delta_Kd_Put2008'], weights=np.ones_like(MM1_2s_All['Delta_Kd_Put2008']) / len(MM1_2s_All['Delta_Kd_Put2008']),
label='pass at 2 sigma')
ax1.hist(MM1_All['Delta_Kd_Put2008'], weights=np.ones_like(MM1_All['Delta_Kd_Put2008']) / len(MM1_All['Delta_Kd_Put2008']) ,
label='pass at 1 sigma', alpha=0.5)
ax1.set_xlabel('ΔKd Putirka 2008')
ax1.set_ylabel('Probability density')
ax1.legend()
# This plots distribution of EnFs with probability on the y axis
ax2.hist(MM1_2s_All['Delta_EnFs_Mollo13'], weights=np.ones_like(MM1_2s_All['Delta_EnFs_Mollo13']) / len(MM1_2s_All['Delta_EnFs_Mollo13']) )#, label='All filters (2sigma)', color='salmon')
ax2.hist(MM1_All['Delta_EnFs_Mollo13'], weights=np.ones_like(MM1_All['Delta_EnFs_Mollo13']) / len(MM1_All['Delta_EnFs_Mollo13']), alpha=0.5)
ax2.set_xlabel('ΔEnFs')
# This plots distribution of DiHd with probability on the y axis
ax3.hist(MM1_2s_All['Delta_DiHd_Mollo13'], weights=np.ones_like(MM1_2s_All['Delta_DiHd_Mollo13']) / len(MM1_2s_All['Delta_DiHd_Mollo13']) )#, label='All filters (2sigma)', color='salmon')
ax3.hist(MM1_All['Delta_DiHd_Mollo13'], weights=np.ones_like(MM1_All['Delta_DiHd_Mollo13']) / len(MM1_All['Delta_DiHd_Mollo13']) , alpha=0.5)
ax3.set_xlabel('ΔDiHd')
ax3.set_ylabel('Probability density')
# This plots distribution of CaTs values with probability on the y axis
ax4.hist(MM1_2s_All['Delta_CaTs_Put1999'], weights=np.ones_like(MM1_2s_All['Delta_CaTs_Put1999']) / len(MM1_2s_All['Delta_CaTs_Put1999']) )#, label='All filters (2sigma)', color='salmon')
ax4.hist(MM1_All['Delta_CaTs_Put1999'], weights=np.ones_like(MM1_All['Delta_CaTs_Put1999']) / len(MM1_All['Delta_CaTs_Put1999']), alpha=0.5)
ax4.set_xlabel('ΔCaTs')
[29]:
Text(0.5, 0, 'ΔCaTs')
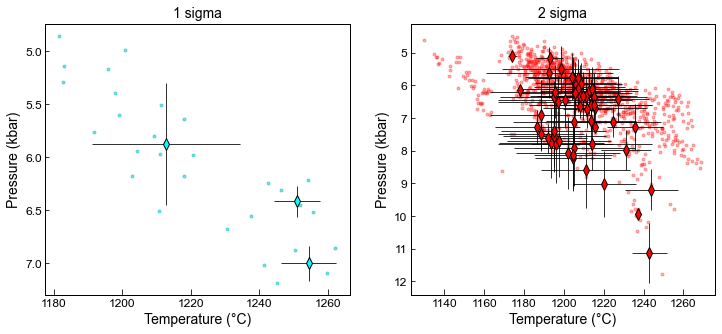
Example 6 - Plotting Pressures and Temperatures
Here comparing first match, with sigma=1 for EnFs, DiHd, CaTs and the second with sigma=2
We show average for cpx with a diamond and error bar, as well as all matches as very faint dots
[30]:
fig, ((ax1, ax2)) = plt.subplots(1, 2, figsize=(12, 5))
# Subplot1
# This plots every Cpx-Liq match, with an opacity of 0.5
ax1.plot(MM1_All['T_K_calc']-273.15, MM1_All['P_kbar_calc'], '.c', alpha=0.5)
# This plots a diamond and error bar for each Cpx averaged with as many liquids as it matches too
ax1.errorbar(MM1_Av['Mean_T_K_calc']-273.15, MM1_Av['Mean_P_kbar_calc'],
xerr=MM1_Av['Std_T_K_calc'].fillna(0), yerr=MM1_Av['Std_P_kbar_calc'].fillna(0),
fmt='d', ecolor='k', elinewidth=0.8, mfc='cyan', ms=8, mec='k')
ax1.set_xlabel('Temperature (°C)')
ax1.set_ylabel('Pressure (kbar)')
ax1.set_title('1 sigma')
# Subplot2 - As above, butmatches with less stringent equilibrium filters
ax2.plot(MM1_2s_All['T_K_calc']-273.15, MM1_2s_All['P_kbar_calc'], '.r', alpha=0.3)
ax2.errorbar(MM1_2s_Av['Mean_T_K_calc']-273.15, MM1_2s_Av['Mean_P_kbar_calc'],
xerr=MM1_2s_Av['Std_T_K_calc'].fillna(0), yerr=MM1_2s_Av['Std_P_kbar_calc'].fillna(0),
fmt='d', ecolor='k', elinewidth=0.8, mfc='red', ms=8, mec='k')
ax2.set_xlabel('Temperature (°C)')
ax2.set_ylabel('Pressure (kbar)')
ax2.set_title('2 sigma')
ax1.invert_yaxis()
ax2.invert_yaxis()
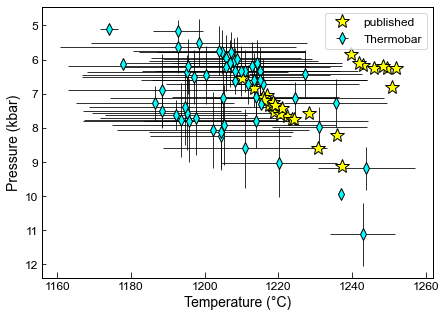
Here we compare our matches to the published data for Gleeson et al. (2020)
The differences result from the fact that the R algorithm of D. Neave throws out matches before T and P are fully converged, and Kd is being calculated using Putirka (1996) in that code, rather than the user-selected thermometer
[31]:
fig, (ax1) = plt.subplots(1, figsize=(7, 5))
# Subplot1
ax1.errorbar(MM1_2s_Av['Mean_T_K_calc']-273.15, MM1_2s_Av['Mean_P_kbar_calc'],
xerr=MM1_2s_Av['Std_T_K_calc'].fillna(0), yerr=MM1_2s_Av['Std_P_kbar_calc'].fillna(0),
fmt='d', ecolor='k', elinewidth=0.8, mfc='cyan', ms=8, mec='k', label='Thermobar')
ax1.plot(Published['Temperature'], Published['Pressure'], '*k', markerfacecolor='yellow', markersize=15, label='published')
ax1.legend()
ax1.invert_yaxis()
ax1.set_xlabel('Temperature (°C)')
ax1.set_ylabel('Pressure (kbar)')
[31]:
Text(0, 0.5, 'Pressure (kbar)')