This page was generated from
docs/Examples/Error_propagation/Cpx_only_contour_plot.ipynb.
Interactive online version:
.
Error propagation - Cpx-only
This notebook gives a worked example showing how to propagate uncertainty for Cpx-only thermometry for different amounts of EPMA-based error on Na\(_2\)O measurements.
We use the functionality provided by Pyrolite to contour the results from Monte Carlo simulations, as well as hexplot.
If you use the pyroplot.density command, please remember to cite Pyrolite! (Williams et al., (2020). pyrolite: Python for geochemistry. Journal of Open Source Software, 5(50), 2314, doi: 10.21105/joss.02314)
This builds on from the notebook showing how to consider error in a single phase (Liquid_Thermometry_error_prop.ipynb). We suggest you look at that first, as its simpler when you don’t have to worry about two separate phases
We consider an example Cpx composition from Gleeson et al. (2020) - https://doi.org/10.1093/petrology/egaa094
You can download the Excel spreadsheet here: https://github.com/PennyWieser/Thermobar/blob/main/docs/Examples/Error_propagation/Gleeson2020_Cpx_Comps.xlsx
You need to install Thermobar once on your machine, if you haven’t done this yet, uncomment the line below (remove the #)
[1]:
#!pip install Thermobar
You will also need to install pyrolite for this example, if you haven’t done so already, uncomment the line below
[2]:
#!pip install pyrolite
[3]:
# Import other python stuff
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import Thermobar as pt
import sympy as sym
pd.options.display.max_columns = None
import pyrolite
Importing data
Note, we haven’t bothered to add the “_Cpx” names after each oxide, so we simply use “suffix=”_Cpx” when we load the data
[4]:
out=pt.import_excel('Gleeson2020_Cpx_Comps.xlsx', sheet_name="Sheet1", suffix="_Cpx")
my_input=out['my_input']
myCpxs1=out['Cpxs']
Lets select 1 Cpx to run the calculations on
The [[ ]] keeps it as a dataframe
[5]:
myCpxs1.iloc[[0]]
[5]:
| SiO2_Cpx | TiO2_Cpx | Al2O3_Cpx | FeOt_Cpx | MnO_Cpx | MgO_Cpx | CaO_Cpx | Na2O_Cpx | K2O_Cpx | Cr2O3_Cpx | Sample_ID_Cpx | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 51.3863 | 0.467 | 4.1961 | 3.8654 | 0.12 | 16.3137 | 21.3039 | 0.378 | 0.0007 | 1.0191 | 0 |
Work out errors
The Cameca software used to analyse these Cpxs returns the error as absolute elemental wt%, ignoring Oxygen, for each element (helpful!). These are loaded in as Na_Err, Si_Err….
This means that you need to do some steps to work out the analytical error on each Cpx. We show these steps below.
First, we decide it easiest to use the convert_oxide_percent_to_element_weight_percent function to convert measured Cpx compositions as oxides into wt% to compare to these errors. Without oxygen = True, does the calculations without oxygen. You can use this function for any phase, just specify what suffix your phase has.
[6]:
Cpx_conv=pt.convert_oxide_percent_to_element_weight_percent(df=myCpxs1, suffix='_Cpx', without_oxygen=True)
Cpx_conv.head()
[6]:
| Si_wt_noO2 | Mg_wt_noO2 | Fe_wt_noO2 | Ca_wt_noO2 | Al_wt_noO2 | Na_wt_noO2 | K_wt_noO2 | Mn_wt_noO2 | Ti_wt_noO2 | Cr_wt_noO2 | P_wt_noO2 | F_wt_noO2 | H_wt_noO2 | Cl_wt_noO2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 43.155184 | 17.675115 | 5.398069 | 27.354400 | 3.989980 | 0.503819 | 0.001044 | 0.166972 | 0.502666 | 1.252750 | 0.0 | 0.0 | 0.0 | 0.0 |
| 1 | 42.432683 | 17.004438 | 5.468846 | 27.558497 | 4.967044 | 0.507369 | 0.000447 | 0.121440 | 0.625890 | 1.313346 | 0.0 | 0.0 | 0.0 | 0.0 |
| 2 | 42.399838 | 16.694183 | 5.608834 | 27.398388 | 5.209677 | 0.523472 | 0.022141 | 0.141541 | 0.681296 | 1.320631 | 0.0 | 0.0 | 0.0 | 0.0 |
| 3 | 42.649103 | 17.224819 | 5.638033 | 27.496627 | 4.631102 | 0.538772 | 0.004336 | 0.097927 | 0.632681 | 1.086601 | 0.0 | 0.0 | 0.0 | 0.0 |
| 4 | 41.057438 | 15.479289 | 7.324030 | 26.604851 | 6.874709 | 0.702197 | 0.025343 | 0.188588 | 1.005908 | 0.737648 | 0.0 | 0.0 | 0.0 | 0.0 |
Now we calculate the % error for each element. For example, for Na, we take the Cameca error (column Na_Err), and compare it to the concentration we just calculated in wt% for Na (column Na_wt_noO2 in the Cpx_conv dataframe)
[7]:
Perc_Err_Na=100*my_input['Na_Err']/Cpx_conv['Na_wt_noO2']
Perc_Err_Na.head()
[7]:
0 8.534819
1 8.553938
2 8.520041
3 8.315210
4 7.049307
dtype: float64
[8]:
# We now need to re-arrange these into a form that we can load into the function.
# We make a new dataframe of these errors
df_Cpx_Err=pd.DataFrame(data={'SiO2_Cpx_Err': 100*my_input['Si_Err']/Cpx_conv['Si_wt_noO2'],
'TiO2_Cpx_Err':100*my_input['Ti_Err']/Cpx_conv['Ti_wt_noO2'],
'Al2O3_Cpx_Err':100*my_input['Al_Err']/Cpx_conv['Al_wt_noO2'],
'FeOt_Cpx_Err':100*my_input['Fe_Err']/Cpx_conv['Fe_wt_noO2'],
'MnO_Cpx_Err':100*my_input['Mn_Err']/Cpx_conv['Mn_wt_noO2'],
'MgO_Cpx_Err':100*my_input['Mg_Err']/Cpx_conv['Mg_wt_noO2'],
'CaO_Cpx_Err':100*my_input['Ca_Err']/Cpx_conv['Ca_wt_noO2'],
'Na2O_Cpx_Err':100*my_input['Na_Err']/Cpx_conv['Na_wt_noO2'],
'Cr2O3_Cpx_Err':100*my_input['Cr_Err']/Cpx_conv['Cr_wt_noO2'],
'K2O_Cpx_Err': 100*my_input['K_Err']/Cpx_conv['K_wt_noO2']}, index=[0])
Lets select 1 Cpx, and its corresponding error as an example
using [[ ]] keeps it as a dataframe, we reset the index because sometimes this can cause python mayhem.
[9]:
Cpx_1=myCpxs1.iloc[[0]].reset_index(drop=True)
Cpx_1_Err=df_Cpx_Err.iloc[[0]].reset_index(drop=True)
Cpx_1_Err
[9]:
| SiO2_Cpx_Err | TiO2_Cpx_Err | Al2O3_Cpx_Err | FeOt_Cpx_Err | MnO_Cpx_Err | MgO_Cpx_Err | CaO_Cpx_Err | Na2O_Cpx_Err | Cr2O3_Cpx_Err | K2O_Cpx_Err | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1.186416 | 2.148544 | 0.907273 | 3.056648 | 27.309951 | 1.129271 | 0.858363 | 8.534819 | 5.172622 | 1934.77739 |
Lets calculate the errors for this Cpx
You could do this for all Cpxs, rather than just this 1 sample, but it’ll take longer, so we do a cut down version here!
20,000 iterations may seem like a lot, but it really helps with the contouring
[10]:
Calc_Cpx_dist=pt.add_noise_sample_1phase(phase_comp=Cpx_1, phase_err=Cpx_1_Err,
phase_err_type="Perc", duplicates=20000, err_dist="normal")
All negative numbers replaced with zeros. If you wish to keep these, set positive=False
Lets calculate pressures and temperatures using a wide variety of equations for Cpx-only thermobarometry
This cell takes abit of time, its 20,000 calculations using a wide variety of P and T equations!
[11]:
Calc_PT=pt.calculate_cpx_only_press_all_eqs(cpx_comps=Calc_Cpx_dist)
Calc_PT.head()
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
c:\Users\penny\Box\Postdoc\MyBarometers\Thermobar_outer\docs\Examples\Error_propagation\Cpx_only_contour_plot.ipynb Cell 21 line 1
----> <a href='vscode-notebook-cell:/c%3A/Users/penny/Box/Postdoc/MyBarometers/Thermobar_outer/docs/Examples/Error_propagation/Cpx_only_contour_plot.ipynb#X26sZmlsZQ%3D%3D?line=0'>1</a> Calc_PT=pt.calculate_cpx_only_press_all_eqs(cpx_comps=Calc_Cpx_dist)
<a href='vscode-notebook-cell:/c%3A/Users/penny/Box/Postdoc/MyBarometers/Thermobar_outer/docs/Examples/Error_propagation/Cpx_only_contour_plot.ipynb#X26sZmlsZQ%3D%3D?line=1'>2</a> Calc_PT.head()
File c:\users\penny\box\postdoc\mybarometers\thermobar_outer\src\Thermobar\clinopyroxene_thermobarometry.py:3365, in calculate_cpx_only_press_all_eqs(cpx_comps, plot, return_cpx, H2O_Liq)
3360 cpx_comps_c['T_Wang21_eq2']=calculate_cpx_only_temp(cpx_comps=cpx_comps_copy, equationT="T_Wang2021_eq2", H2O_Liq=H2O_Liq)
3362 cpx_comps_c['T_Petrelli20']=calculate_cpx_only_temp(cpx_comps=cpx_comps_copy,
3363 equationT="T_Petrelli2020_Cpx_only_onnx")
-> 3365 cpx_comps_c['T_Jorgenson22']=calculate_cpx_only_temp(cpx_comps=cpx_comps_copy,
3366 equationT="T_Jorgenson2022_Cpx_only_onnx")
3368 # cpx_comps_c['T_Petrelli21_H2O']=calculate_cpx_only_temp(cpx_comps=cpx_comps_copy,
3369 # equationT="T_Petrelli2020_Cpx_only_withH2O_onnx").T_K_calc
3370 #
3371 # cpx_comps_c['P_Petrelli21_H2O']=calculate_cpx_only_press(cpx_comps=cpx_comps_copy,
3372 # equationP="P_Petrelli2020_Cpx_only_withH2O").P_kbar_calc
3374 cpx_comps_c['T_Put_Teq32d_Peq32a']=calculate_cpx_only_press_temp(cpx_comps=cpx_comps_copy,
3375 equationP="P_Put2008_eq32a", equationT="T_Put2008_eq32d").T_K_calc
File c:\users\penny\box\postdoc\mybarometers\thermobar_outer\src\Thermobar\clinopyroxene_thermobarometry.py:3537, in calculate_cpx_only_temp(cpx_comps, equationT, P, H2O_Liq, eq_tests)
3533 T_K=df_stats['T_K_calc']
3535 elif ('Petrelli' in equationT or "Jorgenson" in equationT) and "onnx" in equationT:
-> 3537 T_K=func(cpx_comps=cpx_comps_c)
3541 else:
3543 kwargs = {name: cpx_components[name] for name, p in sig.parameters.items()
3544 if p.kind == inspect.Parameter.KEYWORD_ONLY}
File c:\users\penny\box\postdoc\mybarometers\thermobar_outer\src\Thermobar\clinopyroxene_thermobarometry.py:1769, in T_Jorgenson2022_Cpx_only_onnx(P, cpx_comps)
1767 import onnxruntime as rt
1768 path=Path(Thermobar_onnx.__file__).parent
-> 1769 sess = rt.InferenceSession(str(path/"Jorg21_Cpx_only_Temp.onnx"))
1772 input_name = sess.get_inputs()[0].name
1773 label_name = sess.get_outputs()[0].name
File c:\Users\penny\anaconda3\Lib\site-packages\onnxruntime\capi\onnxruntime_inference_collection.py:432, in InferenceSession.__init__(self, path_or_bytes, sess_options, providers, provider_options, **kwargs)
430 raise fallback_error from e
431 # Fallback is disabled. Raise the original error.
--> 432 raise e
File c:\Users\penny\anaconda3\Lib\site-packages\onnxruntime\capi\onnxruntime_inference_collection.py:419, in InferenceSession.__init__(self, path_or_bytes, sess_options, providers, provider_options, **kwargs)
416 disabled_optimizers = kwargs["disabled_optimizers"] if "disabled_optimizers" in kwargs else None
418 try:
--> 419 self._create_inference_session(providers, provider_options, disabled_optimizers)
420 except (ValueError, RuntimeError) as e:
421 if self._enable_fallback:
File c:\Users\penny\anaconda3\Lib\site-packages\onnxruntime\capi\onnxruntime_inference_collection.py:451, in InferenceSession._create_inference_session(self, providers, provider_options, disabled_optimizers)
449 if not providers and len(available_providers) > 1:
450 self.disable_fallback()
--> 451 raise ValueError(
452 f"This ORT build has {available_providers} enabled. "
453 "Since ORT 1.9, you are required to explicitly set "
454 "the providers parameter when instantiating InferenceSession. For example, "
455 f"onnxruntime.InferenceSession(..., providers={available_providers}, ...)"
456 )
458 session_options = self._sess_options if self._sess_options else C.get_default_session_options()
459 if self._model_path:
ValueError: This ORT build has ['AzureExecutionProvider', 'CPUExecutionProvider'] enabled. Since ORT 1.9, you are required to explicitly set the providers parameter when instantiating InferenceSession. For example, onnxruntime.InferenceSession(..., providers=['AzureExecutionProvider', 'CPUExecutionProvider'], ...)
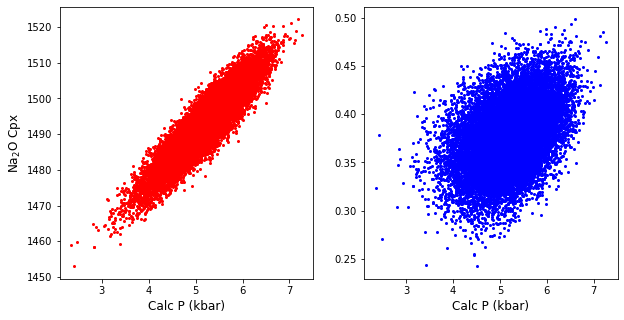
Lets plot these calculations using the Wang et al. (2021) thermometer and barometer
You can see that despite random input errors, P and T end up correlated in the resulting distribution
You can also see that pressure is controlled predominantly by the error on the Na2O component, with scatter around this trend
[ ]:
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(10,5))
ax1.plot(Calc_PT['P_Wang21_eq1'], Calc_PT['T_Wang21_eq2'], '.r' )
ax2.plot(Calc_PT['P_Wang21_eq1'], Calc_PT['Na2O_Cpx'], '.b' )
ax1.set_xlabel('Calc P (kbar)')
ax2.set_xlabel('Calc P (kbar)')
ax1.set_ylabel('Calc T (K)')
ax1.set_ylabel('Na$_2$O Cpx')
Text(0, 0.5, 'Na$_2$O Cpx')
A more robust way to show this
If you play around with duplicates = , you’ll see that the more points you ask for, the more spread in pressure space.
It is more robust to contour results
First, we show how to do this using hexbin (matplotlib)
We underlie the published error on the barometer, at the average calculated composition.
[ ]:
fig, (ax1) = plt.subplots(1, 1, figsize=(6,5))
ax1.hexbin(Calc_PT['P_Wang21_eq1'],
Calc_PT['T_Wang21_eq2'],
cmap=plt.cm.BuPu,
linewidths=0.2, bins=10)
ax1.set_xlabel('Calc P (kbar)')
ax1.set_xlabel('Calc T (K)')
ax1.errorbar(np.mean(Calc_PT['P_Wang21_eq1']),
np.mean(Calc_PT['T_Wang21_eq2']),
xerr=1.52, yerr=35.2,
fmt='.', ecolor='k', elinewidth=0.8, mfc='k', ms=1)
<ErrorbarContainer object of 3 artists>
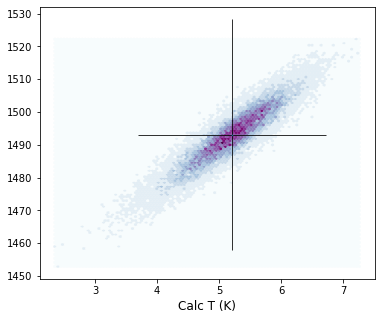
But this is still hard to visualize!
We can add contours using pyrolite, so we can visualize where 67% of simulations lie, and where 95% of simulations lie
You will have to play around a bit with the number of bins, and the extent parameter.
We always plot PT first, then use that to work out the extent, which tells the code where to look for contours in. This only needs to be approximate.
This step can be slow, if you are using 4-8 Gb of Ram, you might want to reduce the number of duplicates when you make the samples.
Remember to cite Pyrolite if you use these contours!
[ ]:
fig, (ax1) = plt.subplots(1, 1, figsize=(6,5))
# This parameter helps pyrolite converge on the contour positions faster,
# e.g. saying look for contours between 3.5 and 7 kbar
# and between 1470 and 1540 K. You'll need to change this for your particular system
extent1=(3.5, 7, 1470, 1540)
ax1.hexbin(Calc_PT['P_Wang21_eq1'],
Calc_PT['T_Wang21_eq2'],
cmap=plt.cm.BuPu,
linewidths=0.2, bins=10)
ax1.set_xlabel('Calc P (kbar)')
ax1.set_xlabel('Calc T (K)')
Calc_PT.loc[:,
["P_Wang21_eq1", "T_Wang21_eq2"]].pyroplot.density(ax=ax1, contours=[0.67, 0.95], colors=['m', 'c'],
extent=extent1, cmap="viridis",
bins=30, label_contours=False)
# Adjust axes limits
ax1.set_xlim([3.5, 7.1])
ax1.set_ylim([1450, 1520])
ax1.errorbar(np.mean(Calc_PT['P_Wang21_eq1']),
np.mean(Calc_PT['T_Wang21_eq2']),
xerr=1.52, yerr=35.2,
fmt='.', ecolor='k', elinewidth=0.8, mfc='k', ms=1)
ax1.set_xlabel('Calc P (kbar)')
ax1.set_ylabel('Calc T (K)')
Text(0, 0.5, 'Calc T (K)')
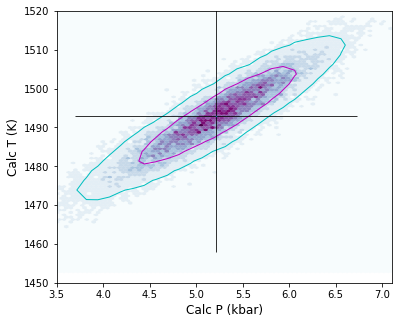
Lets work out what would happen if we doubled the analytical error on Na2O
Be patient, this takes a bit of time, its 20,000 calculations!
[ ]:
# First, lets duplicate the error dataframe
df_Cpx_Err_2Na=df_Cpx_Err.copy()
# Lets set Na2O error to 2* what it was
df_Cpx_Err_2Na['Na2O_Cpx_Err']=2*df_Cpx_Err['Na2O_Cpx_Err']
# And lets take sample 0 again
Cpx_1_Err_2Na=df_Cpx_Err_2Na.iloc[[0]].reset_index(drop=True)
# Lets simulate some new Cpxs.
Calc_Cpx_dist_2Na=pt.add_noise_sample_1phase(phase_comp=Cpx_1, phase_err=Cpx_1_Err_2Na,
phase_err_type="Perc", duplicates=20000, err_dist="normal")
# And lets calculate pressures
Calc_PT_2Na=pt.calculate_cpx_only_press_all_eqs(cpx_comps=Calc_Cpx_dist_2Na)
All negative numbers replaced with zeros. If you wish to keep these, set positive=False
Lets plot these new errors vs. the old errors
Again, the contours can take ~30s to appear (with 16 Gb of RAM)
[ ]:
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(10,5), sharex=True, sharey=True)
extent1=(3.5, 7, 1470, 1540)
ax1.hexbin(Calc_PT['P_Wang21_eq1'],
Calc_PT['T_Wang21_eq2'],
cmap=plt.cm.BuPu,
linewidths=0.2, bins=10)
ax1.set_xlabel('Calc P (kbar)')
ax1.set_xlabel('Calc T (K)')
extent2=(3.5, 7, 1470, 1540)
ax2.hexbin(Calc_PT_2Na['P_Wang21_eq1'],
Calc_PT_2Na['T_Wang21_eq2'],
cmap=plt.cm.BuPu,
linewidths=0.2, bins=10)
ax2.set_xlabel('Calc P (kbar)')
ax2.set_xlabel('Calc T (K)')
Calc_PT.loc[:,
["P_Wang21_eq1", "T_Wang21_eq2"]].pyroplot.density(ax=ax1, contours=[0.67, 0.95], colors=['m', 'c'],
extent=extent1, cmap="viridis",
bins=70, label_contours=False)
Calc_PT.loc[:,
["P_Wang21_eq1", "T_Wang21_eq2"]].pyroplot.density(ax=ax2, contours=[0.67, 0.95], colors=['m', 'c'],
extent=extent1, cmap="viridis",
bins=70, label_contours=False, width=0.5)
Calc_PT_2Na.loc[:,
["P_Wang21_eq1", "T_Wang21_eq2"]].pyroplot.density(ax=ax2, contours=[0.67, 0.95], colors=['r', 'blue'],
extent=extent2, cmap="viridis",
bins=70, label_contours=False)
ax1.errorbar(np.mean(Calc_PT['P_Wang21_eq1']),
np.mean(Calc_PT['T_Wang21_eq2']),
xerr=1.52, yerr=35.2,
fmt='.', ecolor='k', elinewidth=0.8, mfc='k', ms=1)
ax2.errorbar(np.mean(Calc_PT['P_Wang21_eq1']),
np.mean(Calc_PT['T_Wang21_eq2']),
xerr=1.52, yerr=35.2,
fmt='.', ecolor='k', elinewidth=0.8, mfc='k', ms=1)
ax1.set_xlim([3, 8])
ax1.set_ylim([1470, 1550])
ax1.set_xlabel('Calc P (kbar)')
ax1.set_ylabel('Calc T (K)')
ax2.set_xlabel('Calc P (kbar)')
ax2.set_ylabel('Calc T (K)')
ax2.yaxis.set_tick_params(which='both', labelbottom=True)
C:\Users\penny\anaconda3\lib\site-packages\pyrolite\util\plot\density.py:202: UserWarning: The following kwargs were not used by contour: 'width'
cs = contour(
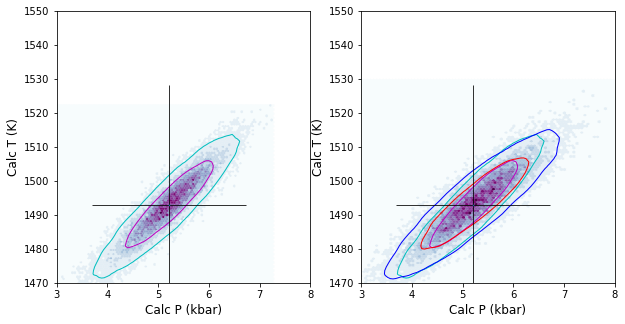
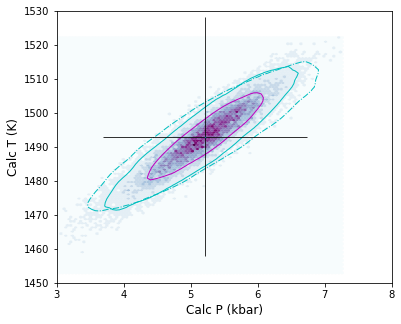
Lets make a pretty figure for publication with just the stated errors for Wang and Putirka thermobarometers, but overlay an additional contour for bigger Na2O errors
[ ]:
# Wang thermobarometers
fig, (ax1) = plt.subplots(1, figsize=(6,5), sharex=True, sharey=True)
## LHS, Wang et al. 2021
extent1=(3.5, 7, 1470, 1540)
ax1.hexbin(Calc_PT['P_Wang21_eq1'],
Calc_PT['T_Wang21_eq2'],
cmap=plt.cm.BuPu,
linewidths=0.2, bins=10)
ax1.set_xlabel('Calc P (kbar)')
ax1.set_xlabel('Calc T (K)')
Calc_PT.loc[:,
["P_Wang21_eq1", "T_Wang21_eq2"]].pyroplot.density(ax=ax1, contours=[0.67, 0.95], colors=['m', 'c'],
extent=extent1, cmap="viridis",
bins=70, label_contours=False)
Calc_PT_2Na.loc[:,
["P_Wang21_eq1", "T_Wang21_eq2"]].pyroplot.density(ax=ax1, contours=[0.95], colors=['c'],
extent=extent2, cmap="viridis",
bins=70, label_contours=False,
linestyles=['-.'])
ax1.errorbar(np.mean(Calc_PT['P_Wang21_eq1']),
np.mean(Calc_PT['T_Wang21_eq2']),
xerr=1.52, yerr=35.2,
fmt='.', ecolor='k', elinewidth=0.8, mfc='k', ms=1)
ax1.set_xlim([3, 8])
ax1.set_ylim([1450, 1530])
ax1.set_xlabel('Calc P (kbar)')
ax1.set_ylabel('Calc T (K)')
ax2.set_xlabel('Calc P (kbar)')
ax2.set_ylabel('Calc T (K)')
ax2.yaxis.set_tick_params(which='both', labelbottom=True)
fig.savefig('Cpx_only_Error.png', dpi=200)
[ ]:
# Putirka Cpx-only barometers
fig, (ax1) = plt.subplots(1, figsize=(6,5), sharex=True, sharey=True)
## LHS, Wang et al. 2021
extent1=(1, 10, 1430, 1540)
ax1.hexbin(Calc_PT['P_Put_Teq32d_Peq32b'],
Calc_PT['T_Put_Teq32d_Peq32b'],
cmap=plt.cm.BuPu,
linewidths=0.2, bins=10)
ax1.set_xlabel('Calc P (kbar)')
ax1.set_xlabel('Calc T (K)')
Calc_PT.loc[:,
["P_Put_Teq32d_Peq32b", "T_Put_Teq32d_Peq32b"]].pyroplot.density(ax=ax1, contours=[0.67, 0.95], colors=['m', 'c'],
extent=extent1, cmap="viridis",
bins=70, label_contours=False)
extent2=(1, 10, 1430, 1540)
Calc_PT_2Na.loc[:,
["P_Put_Teq32d_Peq32b", "T_Put_Teq32d_Peq32b"]].pyroplot.density(ax=ax1, contours=[0.95], colors=['c'],
extent=extent2, cmap="viridis",
bins=70, label_contours=False,
linestyles=['-.'])
ax1.errorbar(np.mean(Calc_PT['P_Put_Teq32d_Peq32b']),
np.mean(Calc_PT['T_Put_Teq32d_Peq32b']),
xerr=2.6, yerr=58,
fmt='.', ecolor='k', elinewidth=0.8, mfc='k', ms=1)
ax1.set_xlim([1.5, 8])
ax1.set_ylim([1430, 1520])
ax1.set_xlabel('Calc P (kbar)')
ax1.set_ylabel('Calc T (K)')
ax2.set_xlabel('Calc P (kbar)')
ax2.set_ylabel('Calc T (K)')
ax2.yaxis.set_tick_params(which='both', labelbottom=True)
fig.savefig('Cpx_only_Error_Put.png', dpi=200)
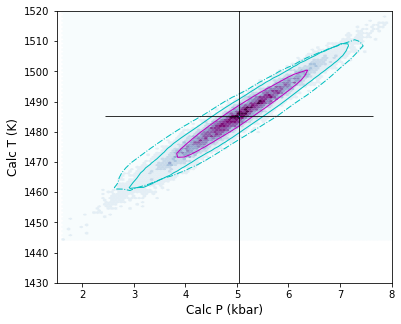
Lets calculate the statistics for various noise simulations
[ ]:
Stats_P=pt.av_noise_samples_series(Calc_PT['P_Wang21_eq1'], Calc_PT['Sample_ID_Cpx_Num'])
Stats_P
| Sample | Mean_calc | Median_calc | St_dev_calc | Max_calc | Min_calc | |
|---|---|---|---|---|---|---|
| 0 | 0.0 | 5.212426 | 5.237335 | 0.576768 | 7.262507 | 2.351651 |
[ ]:
Stats_T=pt.av_noise_samples_series(Calc_PT['T_Wang21_eq2'], Calc_PT['Sample_ID_Cpx_Num'])
Stats_T
| Sample | Mean_calc | Median_calc | St_dev_calc | Max_calc | Min_calc | |
|---|---|---|---|---|---|---|
| 0 | 0.0 | 1493.025062 | 1493.198869 | 8.540826 | 1522.1718 | 1452.932404 |
[ ]:
Stats_T=pt.av_noise_samples_series(Calc_PT_2Na['T_Wang21_eq2'], Calc_PT_2Na['Sample_ID_Cpx_Num'])
Stats_T
| Sample | Mean_calc | Median_calc | St_dev_calc | Max_calc | Min_calc | |
|---|---|---|---|---|---|---|
| 0 | 0.0 | 1493.363786 | 1493.548499 | 9.11549 | 1529.575166 | 1456.638437 |
[ ]:
Stats_P=pt.av_noise_samples_series(Calc_PT_2Na['P_Wang21_eq1'], Calc_PT_2Na['Sample_ID_Cpx_Num'])
Stats_P
| Sample | Mean_calc | Median_calc | St_dev_calc | Max_calc | Min_calc | |
|---|---|---|---|---|---|---|
| 0 | 0.0 | 5.2346 | 5.242048 | 0.714344 | 8.348103 | 2.126887 |
[ ]:
Stats_P_Put=pt.av_noise_samples_series(Calc_PT_2Na['P_Put_Teq32d_Peq32b'], Calc_PT_2Na['Sample_ID_Cpx_Num'])
Stats_P_Put
| Sample | Mean_calc | Median_calc | St_dev_calc | Max_calc | Min_calc | |
|---|---|---|---|---|---|---|
| 0 | 0.0 | 5.013051 | 5.022533 | 0.944134 | 8.845332 | 0.948181 |
[ ]:
Stats_P_Put=pt.av_noise_samples_series(Calc_PT['P_Put_Teq32d_Peq32b'], Calc_PT['Sample_ID_Cpx_Num'])
Stats_P_Put
| Sample | Mean_calc | Median_calc | St_dev_calc | Max_calc | Min_calc | |
|---|---|---|---|---|---|---|
| 0 | 0.0 | 5.019986 | 5.025435 | 0.83673 | 8.116579 | 1.594801 |
[ ]:
Stats_P_Put=pt.av_noise_samples_series(Calc_PT['T_Put_Teq32d_Peq32b'], Calc_PT['Sample_ID_Cpx_Num'])
Stats_P_Put
| Sample | Mean_calc | Median_calc | St_dev_calc | Max_calc | Min_calc | |
|---|---|---|---|---|---|---|
| 0 | 0.0 | 1485.050547 | 1485.152212 | 9.505677 | 1518.411815 | 1446.638034 |
[ ]: